Mechanotransduction
We are determining how cells convert mechanical cues into signals that regulate cell proliferation, differentiation and tissue functions. Studies investigate mechanotransduction across length scales, from atomistic simulations to 3D organoids. We develop engineering and biophysical tools to determine how mechanical stimuli in vivo such as cyclic stretch in lungs regulate tissue functions. Research focuses primarily on intercellular adhesion proteins cadherins, which are critical mechanical and signaling hubs in all tissues. We further identify the clinical significance of force transduction in vascular disease and breast cancer and exploiting these findings to regulate tissue functions. We collaborate with clinicians and cell biologists at UCSF, Univ of Washington, Northwestern, Univ of Toronto, UI Chicago, and UIUC.
Molecular force transduction
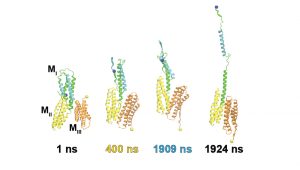
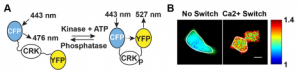
MD simulations, dynamic fluorescence imaging using protein biosensors, and biochemical approaches are identifying molecular mechanisms of force transduction in living cells. Simulations reveal how force alters protein conformations (and function) to activate intracellular signals (force transduction). We test model predictions using live cell fluorescence imaging and cell engineering. Dynamic fluorescence imaging, quantitative immunofluorescence and cell engineering reveal force activated molecular cascades in live cells. Further studies assess the impact of these molecular scale events on cell and tissue functions.