What is ChIPWig?
ChIPWig is a software tool to compress Chromatin immunoprecipitation-sequencing (ChIP-seq) Wig files.
Why ChIPWig?
ChIP-seq is a technique that enables analyzing the interactions between protein and DNA using next generation sequencing technologies. ChIP-seq is an inexpensive DNA sequencing technique which combines ChIP with enormously parallel DNA sequencing to identify the binding sites of DNA-associated proteins. Specifying protein-DNA interactions and their role in regulating gene expression is of crucial importance in many biomedical applications. However, the vast volume of ChIP-seq data has caused challenges in terms of data storage, data transfer and exchange and hence increased the overall cost of data maintenance. To tackle this problem, different data compression techniques have been proposed to reduce the size of ChIP-seq files.
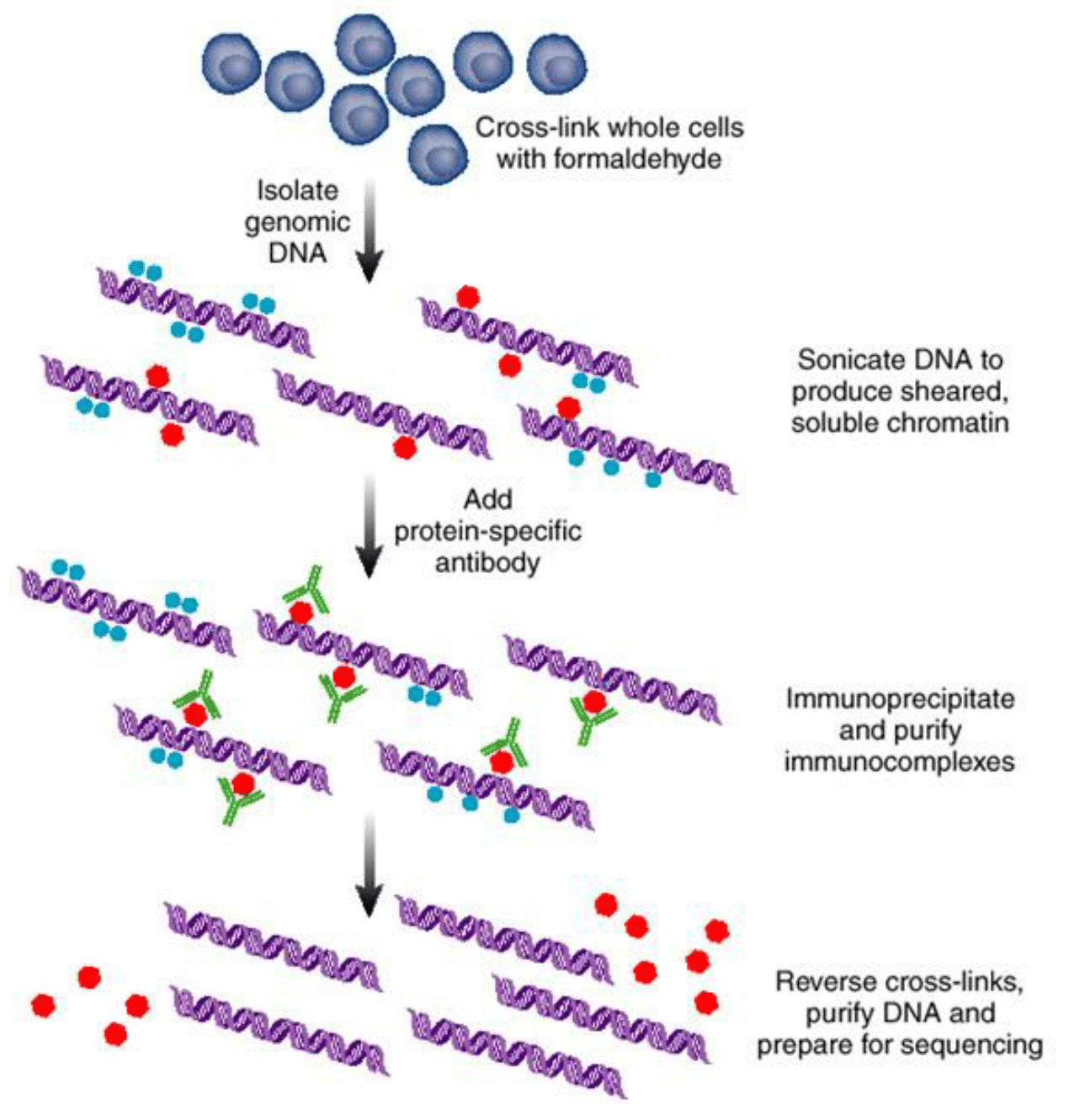
Image credit: Elaine R Mardis ER (2007) ChIP-seq: welcome to the new frontier Nature Methods 4:613–614.
We propose a new low-rate compression method especially designed for ChIP-seq data., termed ChIPWig. Our approach is based on source coding techniques which include transform coding, delta coding, run-length coding and arithmetic coding for integers and correlated real numbers.
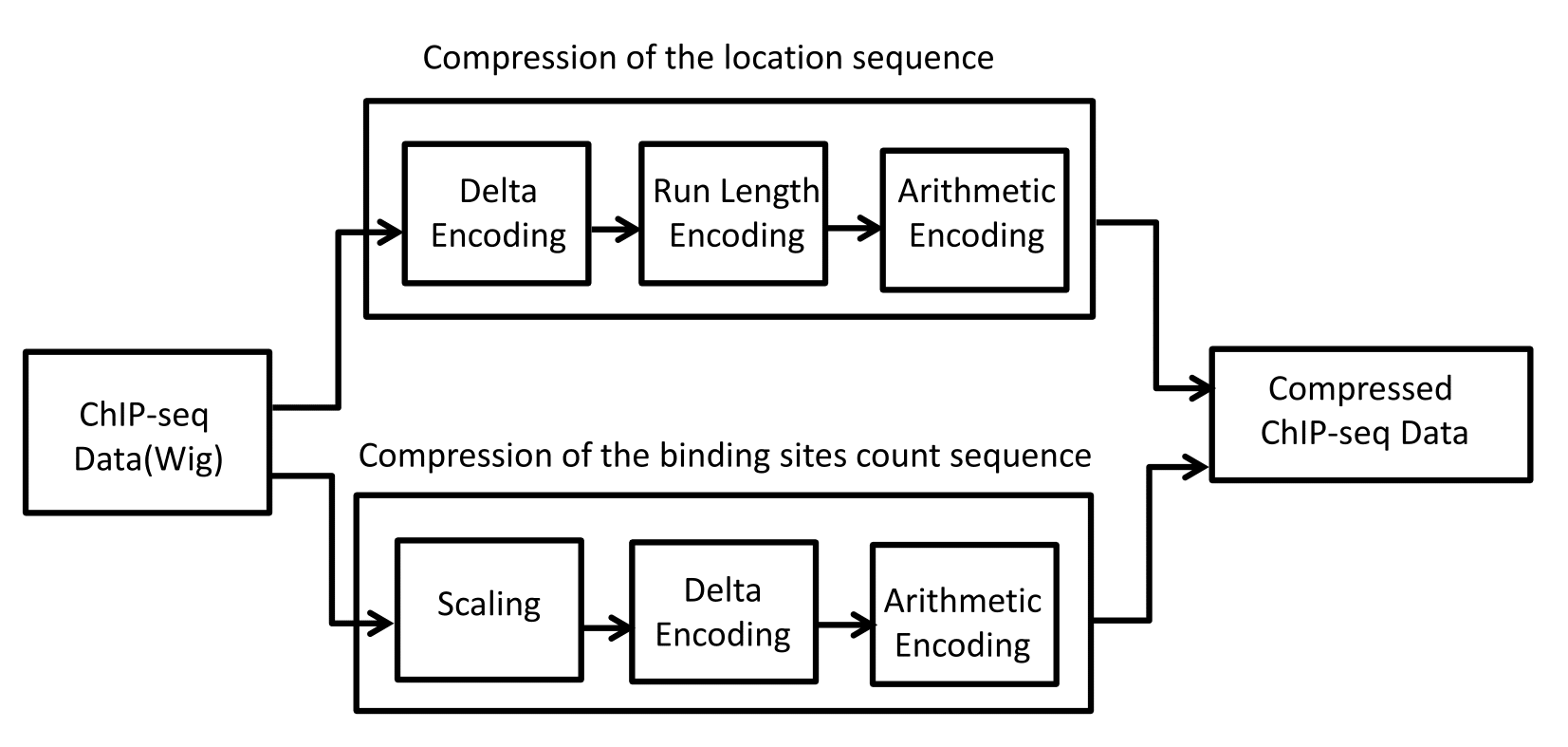
The block model for the ChIPWig compression algorithm.
ChIPWig offers significantly better compression rates than standard bigWig and cWig methods. ChIPWig also offers random access functionalities which enable fast queries from the compressed file. We tested our software for 10 ChIP-seq Wig files from the ENCODE project. The compression rate and running time both in standard mode and random query mode are shown in the following figures.
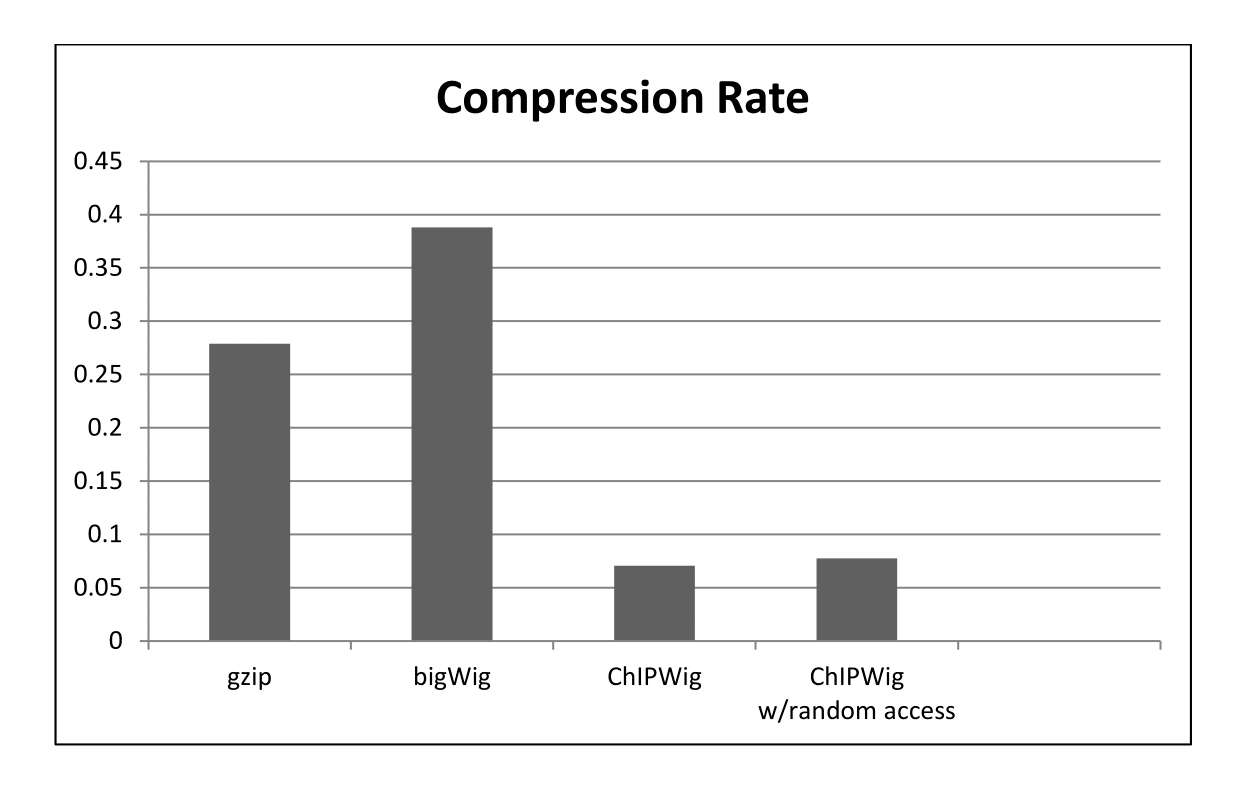
Average Compression Rate of ChIPWig on ENCODE data files Compared to bigWig and gzip.
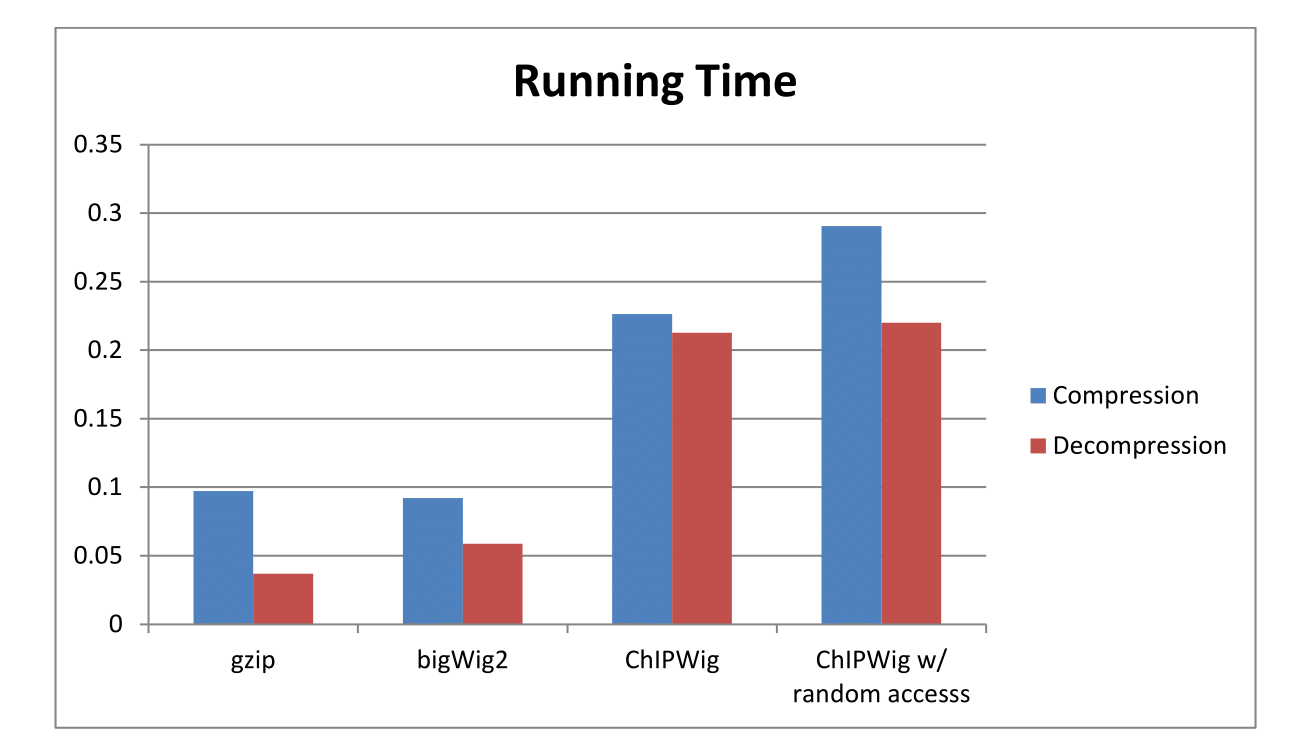
Average Running Time of ChIPWig on ENCODE data files Compared to bigWig and gzip.
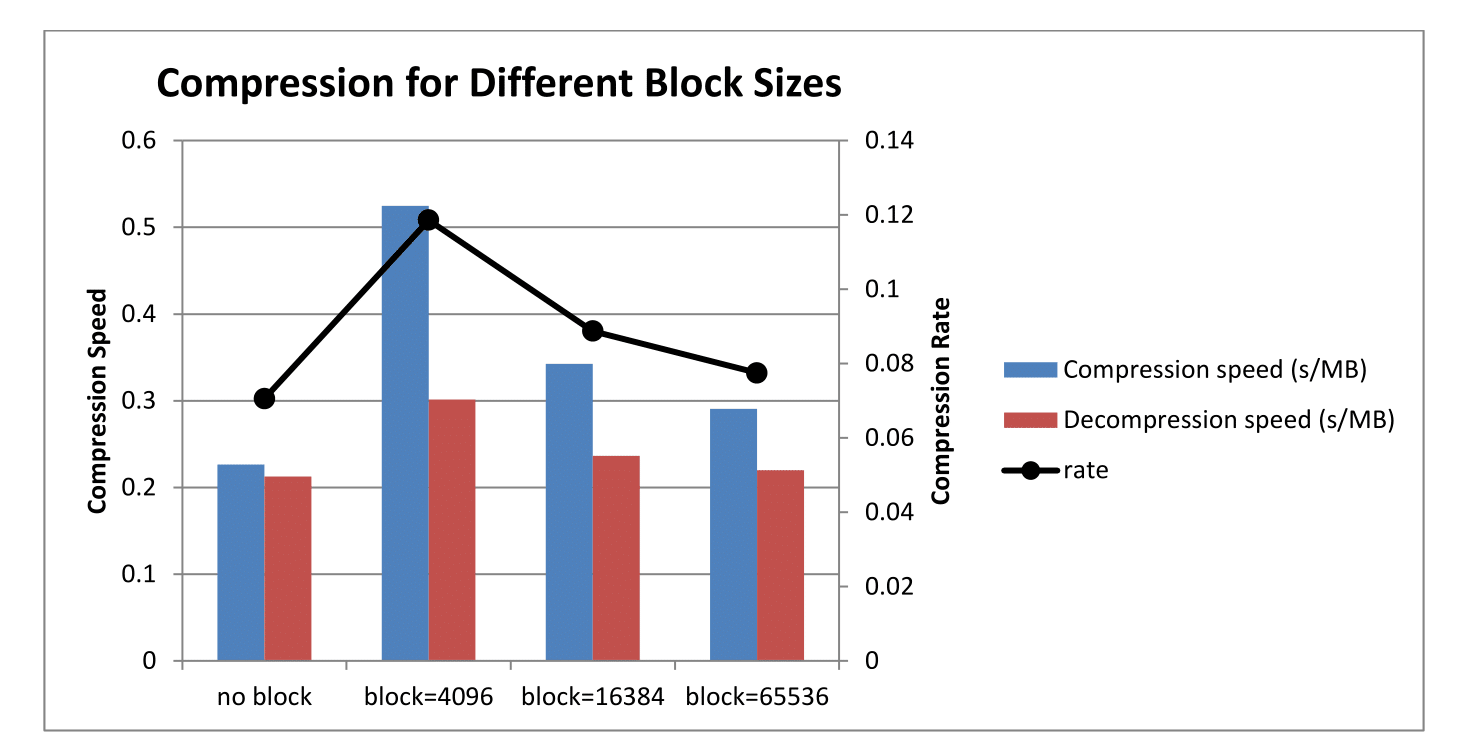
Average Compression Rate of ChIPWig on ENCODE data files Compared to with different block sizes.
How do I download ChIPWig?
You may access ChIPWig’s source code and README file here.
Usage
After following the README file, use the following commands to run ChIPWig.
1. To compress a file, please use:
wig2chipwig [InputFile] [OutputFile]
options:
-r [B, encode block size from 11 to 18] random access and encode by blocks of size 2^B
2. To decompress a file, please use:
chipwig2wig [InputFile] [OutputFile]
options:
-b decompresses a whole file that has been compressed by encoding block-wise.
-s [ChrmName (e.g. chr1)] [Query Start (integer)] [Query End (integer)] subsequence query
Example
1. Compress a file using standard setup:
$ wig2chipwig in.wig out.chipwig
2. Compress a file allowing random query in the future:
$ wig2chipwig in.wig out.chipwig -r 16
3. Decompress a whole file:
$ chipwig2wig in.chipwig out.wig
4. Decompress a whole file that has been compressed by encoding block-wise:
$ chipwig2wig in.chipwig out.wig -b
5.Decompress a file only in chr1, start from location 10001, end at location 11051:
$ chipwig2wig in.chipwig out.wig -s chr1 10001 11051
Notes
1. If your wig file contains numerical values with more than three decimal digits, there may be rare rounding errors. No rounding errors occur during decompression when the precision of the values is less than four digits.
2. Some wig files may appear in single column format in which case it is recommended to add a regular (or custom) location column.
Contact
Olgica Milenkovic ( milenkov@illinois.edu), Vida Ravanmehr (vidarm@illinois.edu)