Abstract: “When antibiotics were introduced in the early twentieth century, they were very effective in treating bacterial infections. However, due to bacterial evolution, misuse and overreliance, these primary antibiotics are becoming less effective. Antibiotic resistance is becoming a major global issue, forcing us to question the stability of our pathogenic defenses. Natural products play a critical role in the development of new antibiotics and provide molecular templates for developing novel classes. Anaephenes A, B, and C are series of alkylphenols, that have shown to be effective antibiotics against Bacillus cereus and Staphylococcus aureus. They were isolated from a marine bacterium off the coast of Guam, known as Hormoscilla oscillatoriales. We have successfully synthesized anaephenes A and B, both which have never been synthesized before. We have confirmed the structure of these molecules using spectroscopic techniques such as 1H and 13C NMR, HMBC, HSQC, and COSY spectra. We have also confirmed their biological activity. Additionally, Anaephenes A and B displayed antimicrobial activity against methicillin-resistant Staphylococcus aureus (MRSA) with MIC values of 16 and 8 ug/mL, respectively.”
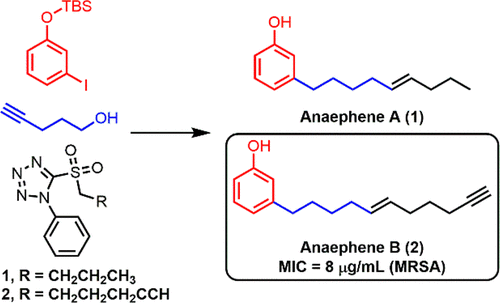


Nice job and clear presentation. Can you possibly address why the yields of the olefination reaction were so low (I calculate ~38%), and strategies to improve these? Additionally, did you see any evidence of isomerization of the terminal alkyne to an internal alkyne in preparing AnaB? Lastly, since you first added an alkynyl chain off the ring, have you considered taking this on and seeing if having it in place vs the CH2CH2 group gives any difference in the activity?
Hello,
Thank you for the questions! I will do my best to answer them all.
1)
For the Julia Olefination, I think the yields were effected the most by human error. There were many opportunities for error in the reaction. For example for adding the aldehyde, it had to be done dropwise at –78 °C then stirred for 1 hour at 0 °C. So maybe I added it too quickly and it reacted with its self, or maybe stirring for 1 hour at 0°C wasn’t long enough. Also, I could have lost some material in the extraction and purification. At the time we only had manual columns for flash chromatography, so thats another opportunity for human error.
Also, we did not synthesize enough of the two pieces to really experiment with the reaction and try to find better conditions, in terms of stir time and solvents for separation. We completed the full route, only going back once to make more of the sulfone and aldehyde.
I also recall having trouble separating the product from biproducts, the RF values were close, I think.
So to get a better yield I would make sure the aldehyde is added slowly and I would experiment with better solvent systems and, if possible , use an automatic column with a gradient setting for the solvent system.
2)
In all honesty my teamate David was in charge of anaB, I of anaA. I don’t recall that happening. But I do know that his Mitsunobu and Julia olefin. reactions were a problem, it was something to do with the alkyne. Hence his yields were slightly lower than mine, which usually isn’t the case. I’ll have to ask him tomorrow
3)
Yes, we knew from the start, that we wanted to keep that alkyne for an analog and we did. For our current project, making the analogs, we’ve made the alkyne version and a version with just the alkyl chain no alkene no alkynes.
The analog with without the alkene and terminal alkyne, but with the alkynyl chain alkyne intact, was the most potent.
Hello,
Thank you for the questions! I will do my best to answer them all.
1)
For the Julia Olefination, I think the yields were effected the most by human error. There were many opportunities for error in the reaction. For example for adding the aldehyde, it had to be done dropwise at –78 °C then stirred for 1 hour at 0 °C. So maybe I added it too quickly and it reacted with its self, or maybe stirring for 1 hour at 0°C wasn’t long enough. Also, I could have lost some material in the extraction and purification. At the time we only had manual columns for flash chromatography, so thats another opportunity for human error.
Also, we did not synthesize enough of the two pieces to really experiment with the reaction and try to find better conditions, in terms of stir time and solvents for separation. We completed the full route, only going back once to make more of the sulfone and aldehyde.
I also recall having trouble separating the product from biproducts, the RF values were close, I think.
So to get a better yield I would make sure the aldehyde is added slowly and I would experiment with better solvent systems and, if possible , use an automatic column with a gradient setting for the solvent system.
2)
In all honesty my teamate David was in charge of anaB, I of anaA. I don’t recall that happening. But I do know that his Mitsunobu and Julia olefin. reactions were a problem, it was something to do with the alkyne. Hence his yields were slightly lower than mine, which usually isn’t the case. I’ll have to ask him tomorrow
3)
Yes, we knew from the start, that we wanted to keep that alkyne for an analog and we did. For our current project, making the analogs, we’ve made the alkyne version
Great talk. How much do we know about the mechanism of action of the anaephenes? Structurally, these compounds are quite small, simple and primarily hydrophobic – what is your strategy for reducing the potential off-target effects? Finally, what do we know about the producing organism? Is Hormoscilla the only genus known to produce this natural product? It would be interesting to know which features of the cyanobacterium mitigate the toxicity of the anaephenes.
Thank you very much.
1)The products where discovered about 2 years ago, so we currently don’t know anything about their mechanism. As organic chemistry enthusiast, we started working towards this doing what we know best. Through organic synthesis and structure activity relationships we can start to make some predictions. We believe the mechanism is involving the bacterial cell wall because of the molecules amphipathic structure. In recent work we’ve seen the compounds perform much better when the amphipathic characteristic is increased. We made analogs without alkynes or alkenes on the chain, and they were much more potent.
2)
We are planning on first, obtaining the most potent analog, and then study the mechanism using microbiology techniques. Afterwards we would make adjustments. But we predict that the mechanism involves the alkyl chain lodging into the cell wall, and then the polar phenol assist in or lyses the cell wall. We hope the mechanism involves peptidoglycan, as the compounds are more effective against gram positive bacteria. If so maybe we can modify the molecule so that it only lyses or effects walls and membranes with peptidoglycan.
3)
I believe Hormoscillia is currently the only known genus to produce these specifically. But there are very similar compounds produced by other marine cyanobacterium. This is from our refrence article, on the discovery and isolation of the anaephene natural products. “The closest relatives to 1–3, the hierridins, were previously reported in species of pico- and filamentous cyanobacteria.24–25 Similarly, the hierridin scaffold contains a polyphenolic head group and a long aliphatic tail. Additional examples of similar natural products are reported from terrestrial plant species including: Spondias tuberosa,26–27 Ozoroa insignis,28 and Piper villiramulum.29–30”
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7315913/
Thank you for your questions!