March 4, 2008
[Download a PDF version]
We are committed to providing the academic community with the most up to date protocols for the expression, purification and characterization of membrane scaffold proteins and the use of these to self-assemble Nanodiscs, both with and without incorporated proteins. All of our procedures have been published, most often in the supplemental materials. The most fundamental are reproduced here.
Materials
Membrane Scaffold Protein: Reconstitute the lyophilized powder using 5 ml of sterile water. The solution will contain MSP in 20 mM Tris (pH 7.4), 0.1 M NaCl, 0.5 mM EDTA, 0.01% NaN3. MSP concentration is determined spectrophotometrically and the concentration is listed on the vial label. Reconstituted MSP containing a 6-His tag can be stored at 4°C for up to several days. For long term storage of solution or lyophilized protein, we recommend temperatures below -20C.
The MSP concentration is determined spectrophotometrically with the molar extinction coefficients depending on the composition of the MSP variant that is used. For MSP1D1 in the standard buffer (20 mM Tris pH 7.4, 0.1 M NaCl, 0.5 mM EDTA, 0.01% NaN3) ε280=21,000 M-1cm-1, for MSP1D1(-), the His-tag cleaved variant, ε280=18,200 M-1cm-1. For MSP1E3D1 ε280=29,400 M-1cm-1, for MSP1E3D1(-) ε280=26,600 M-1cm-1.
Phospholipid: Stocks are prepared in chloroform at 50-100 mM for long-term storage at –20ºC in 4 ml glass vials with Teflon-lined screw caps. The concentration of the stock solution is determined by phosphate analysis (see Determination of Total Phosphorous). Hamilton syringes are used to measure volumes of organic solutions as plastic pipette tips cannot accurately dispense non-aqueous solutions and organic solvents will partially dissolve the plastic tip and contaminate the lipid.
BioBeads SM-2: These polystyrene beads can be purchased from Bio-Rad. The beads should be washed with methanol in advance and thoroughly rinsed with water. BioBeads are stored in water. Alternatively, Amberlite XAD-2 (Sigma catalog number 20275 or 10357 can be used).
Preparation Of Nanodisc Reconstitution Mixtures
Dispense the desired amount of chloroform lipid stock into a disposable glass culture tube. Dry up the solvent using a gentle stream of nitrogen gas in a fume hood. A thin film on the lower walls of the tube can be obtained by rotating the tube while holding it at an angle. To remove residual solvent, place the tube in a vacuum dessicator under high vacuum for at least 4 hr (usually overnight). Dried lipid should have an opaque, white appearance; it should not appear glassy.
Add buffer containing sodium cholate to the dried lipid film (typically cholate is twice the concentration of lipid, e.g. 100 mM cholate in buffer is added to give 50 mM final phospholipid concentration). Vortex the tube, heat it under hot tap water, and sonicate in an ultrasonic bath until the solution is clear, and no lipid remains on the walls of the tube. This is the working lipid stock for the assembly mixture.
Calculate the volumes of detergent solubilized lipid, MSP, buffer and target (if applicable) necessary for the reconstitution mixture. Add MSP solution to cholate-solubilized phospholipid to yield desired lipid to protein ratio:
| Optimal ratios for MSP1 | Optimal ratios for MSP1E3 | Incubation temperature | |
| DPPC | 90:1 | 170:1 | 37 ºC |
| DMPC | 80:1 | 150:1 | 25 ºC |
| POPC | 65:1 | 130:1 | 4 ºC |
Make sure the final cholate concentration in the reconstitution mixture is between 12 – 40 mM. Supplement with standard buffer or cholate stock solution if necessary. Incubate the mixture at the appropriate incubation temperature for at least 15 minutes or longer.
Preparation Of Nanodiscs
The self-assembly process is initiated upon removal of the detergent. Cholate can be removed by dialysis (at the incubation temperature) or with BioBeads SM-2 (Bio-Rad).
Dialysis: Dialyze the sample against 3 changes of buffer over 24 hr. The volume of buffer should be ~1000 times the volume of MSP/lipid/cholate mixture.
BioBeads: Remove most of the water from the beads immediately before use. Add 0.5-0.8 g of the damp beads per every ml of the reconstitution mixture. Place the suspension on the orbital shaker and incubate for at least 2 h (DPPC, DMPC) and 4 h for POPC.
Nanodisc samples are fractionated on a Superdex 200 10/300 GL column (GE Healthcare) in MSP Standard Buffer, with a flow rate of 0.5 ml/min. The upper limit for injection volume is 0.5 ml. Samples are filtered prior to injection and fractions are collected every minute. A typical chromatogram is shown:
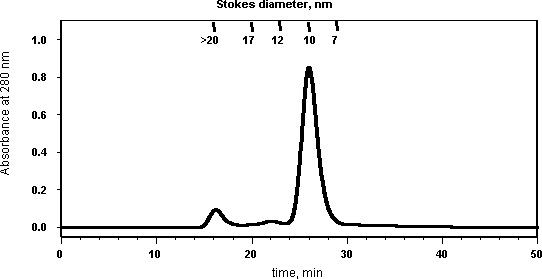  |
| Standards (Stokes diameter in nm): Thyroglobulin (bovine thyroid) (17 nm), Ferritin (horse spleen) (12.2 nm), Catalase (bovine liver) (10.4 nm), Albumin (bovine serum) (7.1). |
References:
1. Bayburt, T. H., Grinkova, Y. V., & Sligar, S. G., 2002, Self-assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins. Nanoletters, 2: 853-856.
2. Denisov, I. G., Grinkova, Y. V., Lazarides, A. A., & Sligar, S. G., 2004, Directed Self-Assembly of Monodisperse Phospholipid Bilayer Nanodiscs with Controlled Size. J. Am. Chem. Soc., 126: 3477-3487.
Guidelines for Incorporation of Membrane Protein Targets into Nanodiscs
Indicated is a typical protocol for incorporating a membrane protein into POPC Nanodiscs
- Prepare disc reconstitution mixture following the protocol for plain discs
- Incubate target protein with secondary detergent
- Mix disc reconstitution mixture with the detergent solubilized target protein (typically we use relatively high MSP to target protein molar ratios), and incubate it on ice for 5 minutes or longer. Make sure that final cholate concentration in the reconstitution mixture exceeds 14 mM
- Add washed BioBeads (0.5-1 g of wet BioBeads per ml), gently shake the mixture using orbital shaker to keep BioBeads suspended for 4-18 hours
Comments
The general idea is quite simple – cholate-solubilized phospholipids are mixed with MSP and detergent solubilized membrane protein, everything is incubated together, then the detergents are removed (usually with BioBeads), and the self-assembly takes place. The important parameters are: (1) lipid to MSP ratio, (2) temperature, (3) choice of detergent and (4) the final lipid and detergents concentrations in the reconstitution mixture. The lipid to MSP ratio depends on the kind of lipid and MSP (it is discussed in details in our papers – Bayburt et al., 2002; Denisov et al., 2004). When relatively high MSP to target protein molar ratios are used, the same lipids to MSP ratio can be used, as for “empty” discs. When the assembly is done at low MSP to target protein ratios, it is likely, that the lipids to MSP ratio will have to be adjusted for optimal assembly (see, for example Bayburt et al, 2006). The temperature should be close to the lipid melting temperature. With POPC we usually conduct the assembly on ice. The lipid and detergent concentrations in the reconstitution mixture should be high enough in order for self-assembly to work properly. The exact value depends on the temperature, cholate to lipid ratio, and on the presence and concentration of the second detergent. We usually keep lipid concentration in the final mixture between 7 mM an 18 mM when cholate to lipid ratio is 2:1; if it is necessary to perform assembly at lower lipid concentration, additional cholate should be introduced into the reconstitution mixture, so the final cholate concentration will be higher than 14 mM.
Note that the choice and concentration of the secondary detergent depend entirely on the membrane protein, and have to be worked out for every new target. In particular, since it is usually not known how many lipids a particular protein will displace in the fully assembled Nanodisc, a titration at various lipid:MSP:target ratios is usually needed. It is important to remember that in the presence of secondary detergent often larger amount of BioBeads is needed (we usually use 0.8-1 g of wet BioBeads per ml of reconstitution mixture, when detergents with low CMC, such as emulgen 913 or triton X-100, are being used). Typically we use excess MSP relative to target protein, and separate discs with incorporated membrane protein from “empty” discs. So far the combination of size-exclusion and affinity chromatography (when tag is on the membrane protein) produced best results. Also note that the membrane protein must be soluble is some detergent that can be removed by dialysis and/or hydrophobic bead treatment. For instance, a soluble aggregated protein will not self-assemble into the Nanodisc. Since the entire procedure can be executed quickly, it is not necessary that the target protein be stable in the detergent for very long periods of time (e.g. that which might be involved in a normal protein purification procedure) but it must be initially solubilized so that the process of self-assembly can ensue. We also have data that suggests that the speed of detergent removal is important in the assembly process, particularly for oligomeric proteins or where it is desired to incorporate a complex of membrane proteins. Thus, the time of detergent removal as modulated by quantity and hydrophobic bead treatment protocol may need to be varied.
Also note that glycerol in relatively high concentrations interferes with the self-assembly. Final glycerol concentration in the disc reconstitution mixture should be below 3%. It is fine, however, to add glycerol to the final disc sample.
Guidelines for Incorporation of Membrane Protein Targets into Nanodiscs
Mix the Nanodisc reconstitution mixture with the membrane protein (solubilized with suitable detergent) at the desired molar ratios. For a relatively pure target protein, we typically use an excess ratio (>4) MSP:target membrane protein. This ratio is dependent on the desired oligomerization state of the protein and the concentration of other membrane proteins in the target preparation. For microsomes or complex mixtures, the total membrane protein concentration should be considered as well since these proteins can also incorporate into discs.
If necessary, adjust the volume of the mixture with Standard Buffer to ensure that the lipid concentration during disc formation is between 5-20 mM). Incubate the assembly mixture for up to 2 hr at the appropriate temperature as determined by the lipid being used. Detergent is typically removed by addition of a excess of BioBeads. Note that triton, emulgen, and similar detergents are known to adsorb very poorly to BioBeads, and thus require an increased excess of beads over the typical amount used for cholate. Assembled Nanodiscs can be purified by affinity chromatography using the 6-His tag on the MSP or a purification method appropriate for the target protein, if available. Exchange buffer prior to affinity chromatography if necessary (e.g. to remove EDTA prior to using a Ni++ affinity column). Fractionate the resulting sample using size-exclusion chromatography.
When selecting a suitable detergent to solubilize the target, usual conditions call for a detergent that results in soluble monomers of most or all of the target protein and allows for recovery of activity after the perturbant is removed. Activity does not necessarily have to be maintained while the protein is detergent solubilized in the assembly mixture. Protease inhibitors should also be included in the reconstitution mixture if protein degradation is a concern. Investigation of different detergent solubilization conditions is an important early experiment when working with target proteins that are being newly tested for incorporation into Nanodiscs. For novel targets, information from studies with vesicles, purification methodologies, and results with related proteins, can provide guidance for detergent selection.
References:
1. Bayburt, T. H., & Sligar, S. G., 2003, Self-assembly of single integral membrane proteins into soluble nanoscale phospholipid bilayers. Prot. Sci., 12: 2476-2481.
2. Civjan, N. R., Bayburt, T. H., Schuler, M. A., & Sligar, S.G., 2003, Direct solubilization of heterologously expressed membrane proteins by incorporation into nanoscale lipid bilayers. BioTechniques, 35: 556-558, 560, 562-563.
3. Sligar, S. G., 2003, Finding a single-molecule solution for membrane proteins. Biochem. Biophys. Res. Comm., 312: 115-119.
4. Baas, B. J., Denisov, I. G., & Sligar, S. G., 2004, Homotropic cooperativity of monomeric cytochrome P450 3A4 in a nanoscale native bilayer environment. Arch. Biochem. Biophys., 430: 218-228.
5. Duan, H., Civjan, N. R., Sligar, S. G., & Schuler, M. A., 2004, Co-incorporation of heterologously expressed Arabidopsis cytochrome P450 and P450 reductase into soluble nanoscale lipid bilayers. Arch. Biochem. Biophys., 424: 141-153.
Determination of Total Phosphorus
This procedure was adapted from the Avanti Polar Lipids website, a good source of practical information about phospholipids and their handling.
Reagents and equipment:
Deionized water
Conc. H2SO4 (Mallinckrodt, No. 2876)
Ammonium molybdate(VI) tetrahydrate (Sigma-Aldrich, A7302)
L-Ascorbic acid (Sigma-Aldrich, No. 25,556-4)
0.65 mM Phosphorus standard solution (Sigma, No. P3869)
Hydrogen peroxide (Fisher, No. H323-500)
A hot plate or sand bath capable of temperatures above 200°C
An aluminum block for test tubes (necessary to maintain and evenly distribute heat)
Disposable glass test tubes (13×100 mm)
Prepare solutions:
8.9 N H2SO4 solution: Slowly add 247 ml of conc. H2SO4 to 753 ml of deionized water, allow the heat to dissipate from the 1L volumetric flask, and mix the solution well. Store the solution in a sealed container at room temperature for up to 1 year.
10% Ascorbic acid solution: Place 10 g of ascorbic acid into a 100 ml volumetric flask. Add 50 ml and mix the solution well. Dilute to 100 ml with deionized water. Store the solution in an amber screw-cap bottle at 4°C for up to 1 month.
2.5% Ammonium molybdate(VI) tetrahydrate solution: Place 2.5 g of ammonium molybdate(VI) tetrahydrate into a 100 ml volumetric flask. Add 50 ml and mix the solution well. Dilute to 100 ml with deionized water. Store the solution in an amber screw-cap bottle at 4°C for up to 1 month.
Prepare sample tubes:
Place sample into the bottom of each tube (Usually 1 and 2 µl in triplicate). Gently remove any solvent from the tubes with N2. Place the following quantities of phosphorus standard into nine separate tubes: 0 nmoles (0 µl) blank, 16.2 nmoles (25 µl), 32.5 nmoles (50 µl), 48.8 nmoles (75 µl), 65.0 nmoles (100 µl), 81.2 nmoles (125 µl), 97.5 nmoles (150 µl), 113.8 nmoles (175 µl), 130.0 nmoles (200 µl), 162.5 nmoles (250 µl).
Digestion of organic sample to inorganic phosphate:
The samples, six standards should be labeled and placed in a vinyl-coated test tube rack.
Add 225 µl H2SO4 to each of the standard tubes and sample tubes. Heat all tubes in an aluminum block in the hood at 200-215°C for 25 min. (Important: temperature must be above 200°C to liberate the phosphate.) Remove tubes from the block and allow them to cool 5 min before adding 75 µl H2O2 to the bottom of all tubes. Replace the tubes in the block and continue to heat for an additional 30 min. The samples should be colorless at this point (if any brown color persists, add 50 µl of H2O2 to all cooled tubes and continue heating tubes for 15 min). Cool tubes to ambient temperature. Add 1.95 ml deionized water to all tubes. Add 0.25 ml ammonium molybdate(VI) tetrahydrate solution to all tubes and vortex each tube to mix. Add 0.25 ml ascorbic acid solution to all tubes and vortex each tube to mix. Heat all tubes at 100°C for 7 min. Cool the tubes to ambient temperature before analysis.
Spectrophotometric analysis of samples:
Zero the spectrophotometer using the 0 nmoles standard (reagent blank). Determine the absorbance of each of the five standards at 820 nm. Determine the absorbance of each of the samples at 820 nm. Generate a calibration curve using the standards and determine the concentration of phosphorus in the samples. The use of a 96 well plate and a microplate capable spectrophotometer is useful to reduce analysis time and take four readings per sample.
The samples should be treated as chemical waste and collected in an appropriately labeled waste container.
References:
Chen, Toribara, and Warner (1956) Anal. Chem. 28:1756-1758.
Fiske & Subbarow (1925) J. Biol. Chem. 66:374-389.
