Both horizontal gene transfer and vertical mutation inheritance contribute to microbial evolution (Arber 2008). Horizontal gene transfer can speed up microbial evolution by preferential selection through environmental stressors, such as, residual antibiotics, heavy metals, as well as other organic compounds (Rensing, Newby et al. 2002; Martinez, Wang et al. 2006; Liang, Wang et al. 2011; Obolski and Hadany 2012). Natural transformation, as one of the main horizontal gene transfer mechanisms, describes the uptake of extracellular DNA into new species. Natural transformation is the only mechanism both DNA and cells are under environmental stresses.
DNA under Environmental Stresses:
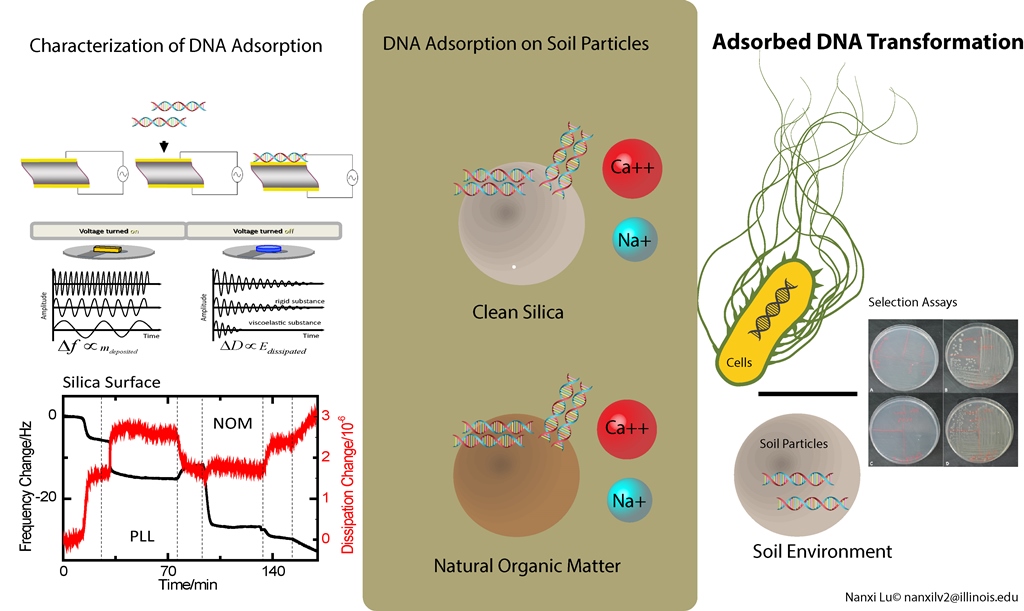
DNA in the environment persists on soil surfaces and re-enters microbial communities by transferring into new species and expressing encoded functions. This study found that DNA adsorbed on soil surfaces maintains the ability to transform into bacterial cells with the presences of different divalent cations and natural organic matters, which influence its conformation and interaction with cells.
Publications:
N. Lu, S. E. Mylon, R. Kong, R. Bhargava, J. L. Zilles, and T. H. Nguyen. 2012. Interactions between dissolved natural organic matter and adsorbed DNA and their effect on natural transformation of Azotobacter vinelandii. Science of the Total Environment, 426: 430-435. Full Tex Link.
N. Lu, J. L. Zilles, and T. H. Nguyen. 2010. Adsorption of extracellular chromosomal DNA and its effects on natural transformation of Azotobacter vinelandii. Applied and Environmental Microbiology, 76(13): 4179-4184. Full Text Link.
Bacterial Cells in the Environment:
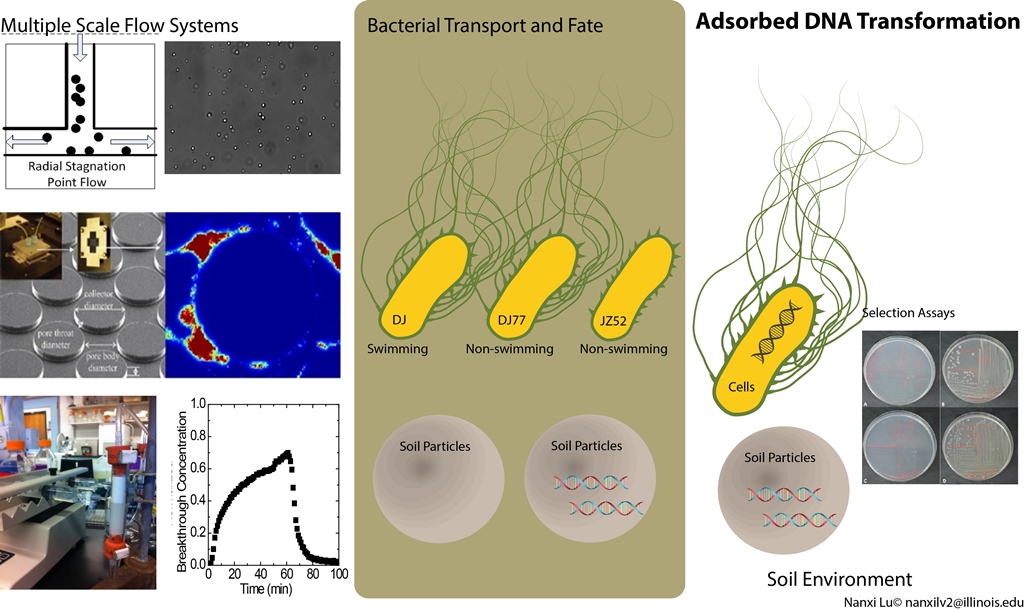
Some bacterial cells are motile in the environment. The swimming ability renders the bacterial cells “space ships searching for strange new worlds and seeking out new microbial life forms”. The DNA carrying new ecological fitness can be one of the targets for this search. This work provides evidences that the swimming ability controls the transport and fate of bacterial cells as well as their interaction with different forms of DNA, dissolved in the soil pore water or adsorbed on soil surfaces.
Publications:
A. Massoudieh, N. Lu, X. Liang, T. H. Nguyen, T. R. Ginn. 2013. Bayesian Process-Identification in Bacteria Transport in Porous Media. Journal of Contaminant Hydrology, 153: 78-91. Full Text Link.
N. Lu, T. Bevard, A. Massoudieh, C. Zhang, A. Dohnalkova, J. L. Zilles, and T. H. Nguyen. 2013. Flagella-mediated differences in deposition dynamics for Azotobacter vinelandii in porous media. Environmental Science & Technology, 47 (10): 5162–5170. Full Text Link.
References:
Arber, W. (2008). “Molecular mechanisms driving Darwinian evolution.” Mathematical and Computer Modelling 47(7–8): 666-674.
Liang, B., G. Wang, et al. (2011). “Facilitation of Bacterial Adaptation to Chlorothalonil-Contaminated Sites by Horizontal Transfer of the Chlorothalonil Hydrolytic Dehalogenase Gene.” Applied and Environmental Microbiology 77(12): 4268-4272.
Martinez, R. J., Y. Wang, et al. (2006). “Horizontal Gene Transfer of PIB-Type ATPases among Bacteria Isolated from Radionuclide- and Metal-Contaminated Subsurface Soils.” Applied and Environmental Microbiology 72(5): 3111-3118.
Obolski, U. and L. Hadany (2012). “Implications of stress-induced genetic variation for minimizing multidrug resistance in bacteria.” BMC Medicine 10(1): 89.
Rensing, C., D. T. Newby, et al. (2002). “The role of selective pressure and selfish DNA in horizontal gene transfer and soil microbial community adaptation.” Soil Biology and Biochemistry 34(3): 285-296.