Research Topic 1 – Nanobubble Therapeutics
We have developed oxygen-encapsulated cellulose nanobubbles and have successfully tested the concept of ultrasound-triggered cargo delivery for treating bladder and prostate cancer (nanobubble size and ultrasound frequency dependent) as shown in Figure 1 by mitigating cancer hypoxia (US Patent 10,670,581, US20160166716A1).
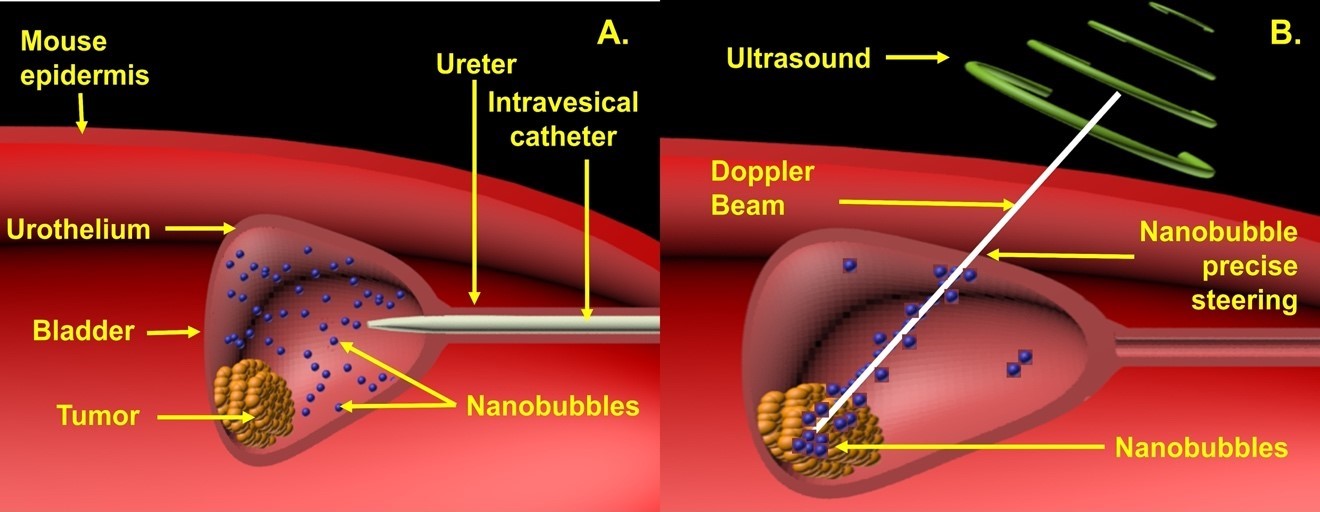 Figure 1. Intravesical treatment in mice bladder cancer. (A) ONBs introduced into the bladder by an intravesical catheter. (B) Precise localization of ONBs (ONBs in blue) to the hypoxic tumor microenvironment using Doppler ultrasound steering with simultaneous B-mode US imaging.
Figure 1. Intravesical treatment in mice bladder cancer. (A) ONBs introduced into the bladder by an intravesical catheter. (B) Precise localization of ONBs (ONBs in blue) to the hypoxic tumor microenvironment using Doppler ultrasound steering with simultaneous B-mode US imaging.
We have subsequently expanded the applications to developing active oxygen-generating nanocarriers and have tested their efficacy in treating pancreatic tumors (Collaborative research with Carle Cancer Center, Carle Foundation Hospital).
We have now reformulated our initial design to address ischemic diseases of the eye, specifically orphan diseases such as Central Retinal Artery Occlusion (CRAO) and Branch Retinal Artery Occlusion (BRAO), as well as Diabetic Retinopathy (Collaborative research Carle Ophthalmology), illustrated in Figure 2. Our goals are to expand our formulation to produce a stable product for utilization in clinical trials.
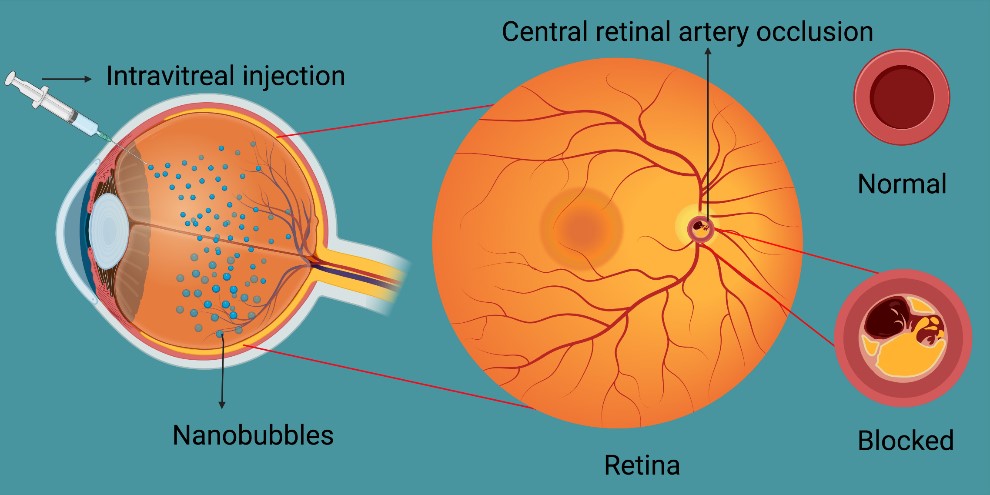 Figure 2. Intravitreal administration of dextran ONBs to treat CRAO
Figure 2. Intravitreal administration of dextran ONBs to treat CRAO
Our oxygen encapsulation concepts are now being translated to developing oxygenated gels to treat chronic wounds, in collaboration with the clinicians at the Wound Healing Center at Carle Foundation Hospital. During the process of technology development, we strive to address several basic mechanisms related to oxygen diffusion in tissues, targeting and uptake by specific cells, biodistribution, clearance, stability, and mechanism of action.
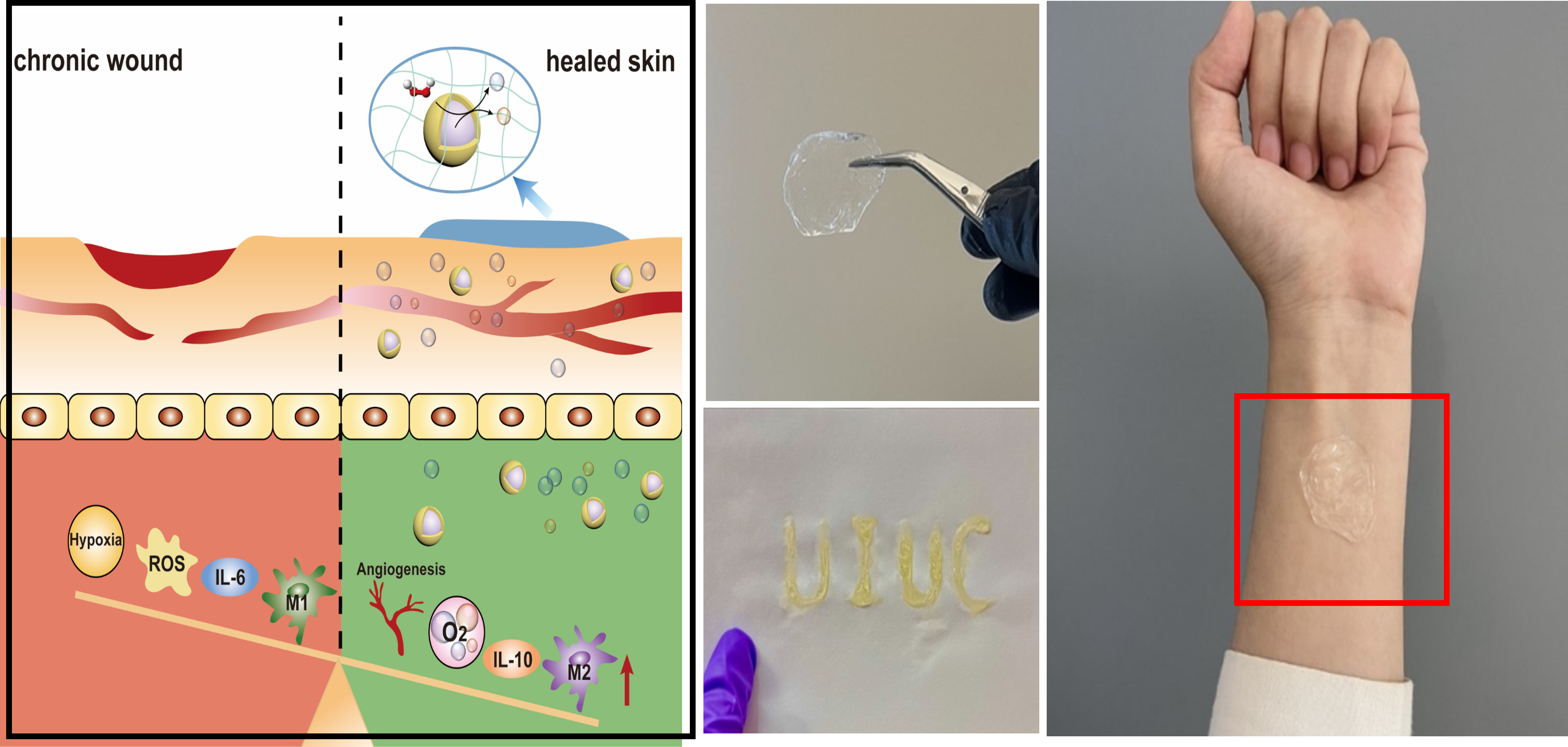 Figure 3. Developing oxygenated gels to treat chronic wounds.
Figure 3. Developing oxygenated gels to treat chronic wounds.
Oxygen Nanobubbles in a Cell
Steering of Oxygen Nanobubbles by ultrasound in the bladder of a tumor bearing mouse
Research Topic 2 – Single molecule super-resolution technologies for dynamic monitoring of epigenetic regulation in relation to nucleus deformation.
The cancer cell nucleus deforms as it invades the interstitial spaces in tissues and the tumor microenvironment. Our focus is to understand the alteration of the chromatin structure in the context of epigenetic regulation and gene expression in a deformed nucleus as cells migrate. We expanded the Fluorescence Correlation Spectroscopy/Fluorescence lifetime imaging bench and developed a super-resolution (Stimulated Emission Depletion-based, STED-based) module to evaluate viscosity, protein diffusion, and chromatin mobility in live cells when cells traverse through constricted spaces (ACS Nano, 2022; Figure 4). We utilized a nanofiber platform (ACS Nano, 2022) to provide a pathway for cells to travel and evaluated the dynamic heterochromatin states in anisotropic nuclei as the nuclei are deformed because cells travel through these constricted tracks created by aligning nanofibers. The next steps will constitute evaluating epigenetic regulation in the context of chromatin diffusion and gene expression due to nucleus stretching upon the application of external forces, and its effect on cell migration.
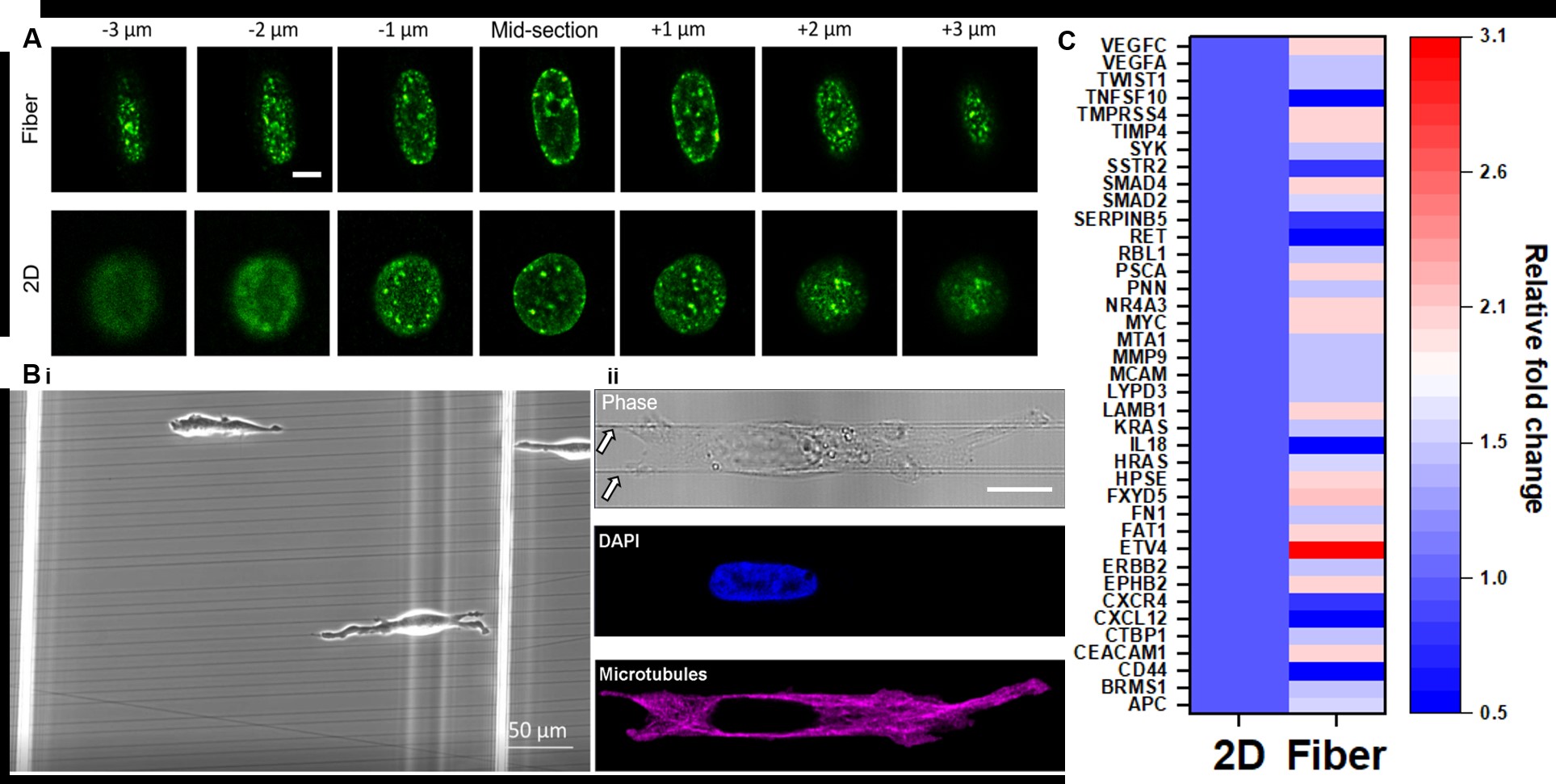 Figure 4. (A) H3K9me3 patterning (z-stack) in stretched nuclei (top row) due to cells migrating on aligned nano-fiber tracks (B) and 2D culture (row 2, A). (C) Relative fold change in gene expression of cells on fibers vs control (2D)
Figure 4. (A) H3K9me3 patterning (z-stack) in stretched nuclei (top row) due to cells migrating on aligned nano-fiber tracks (B) and 2D culture (row 2, A). (C) Relative fold change in gene expression of cells on fibers vs control (2D)
Live cell Kinase signaling visualized with peptide biosensors targeting the kinase domain – Fluorescence Lifetime imaging monitors VEGF kinase signaling
Click here for a snapshot of Single Molecule Florescence Research in our lab
Research Topic 3 – Cancer Toxicology
Our group has intensely focused on exploring epigenetic toxicity of Per- and polyfluoroalkyl substances (PFAS). Along with a team at UIUC in collaboration with external partners, we are exploring the effect of PFAS on the liver, kidney, and intestine. We have identified a set of genes and epigenetic targets triggered by PFAS toxicity. We utilize single-cell techniques, and molecular biology tools along with sequencing and metabolomic analysis to assess the toxicity. Specific projects in this area are:
• Development of epigenome editing tools for toxicity reversal
• Development and utilization of machine learning tools for intelligent prediction of toxicity biomarkers.
• Assess the effect of co-culture systems (microbiome-epigenome)on PFAS toxicity
We collaborate with Cancer biologists to develop generalized platforms to assess toxicity mechanisms. We work very closely with the Illinois EPA on PFAS quantification in aquatics and soil samples. In addition, we are also exploring avenues to assess PFAS levels in firefighters in collaboration with the Illinois Fire Service Institute. More recently, we have incorporated machine learning tools to identify biomarkers related to epigenetic regulation. Details of the broader impact of this study can be found at: isrc.illinois.edu
Research Topic 4 – Single nanoparticle in cell technologies and biosensors
We are the first group to demonstrative live cell quantification of mRNA utilizing the plasmons properties of gold nanoparticles. We have further expanded this work for single cell epigenetic profiling and sensing of molecular markers in different cellular compartments. In the long-term our goal is to further translate our concepts for multicomponent tissue profiling utilizing gold nanoparticle probes, quantum/carbon dots at single molecule/particle resolution. This work will be conducted at the Laboratory in Carle Hospital and Beckman Institute.
Live Cell quantification of mRNA – Gold nanoparticle probes target splice junction of BRCA1 mRNA to form dimers (bright spots) – Nature Nanotechnology (2014)
Click here for a sample of Plasmonic Sensors research to Quantify Molecular Markers in Single Cells
