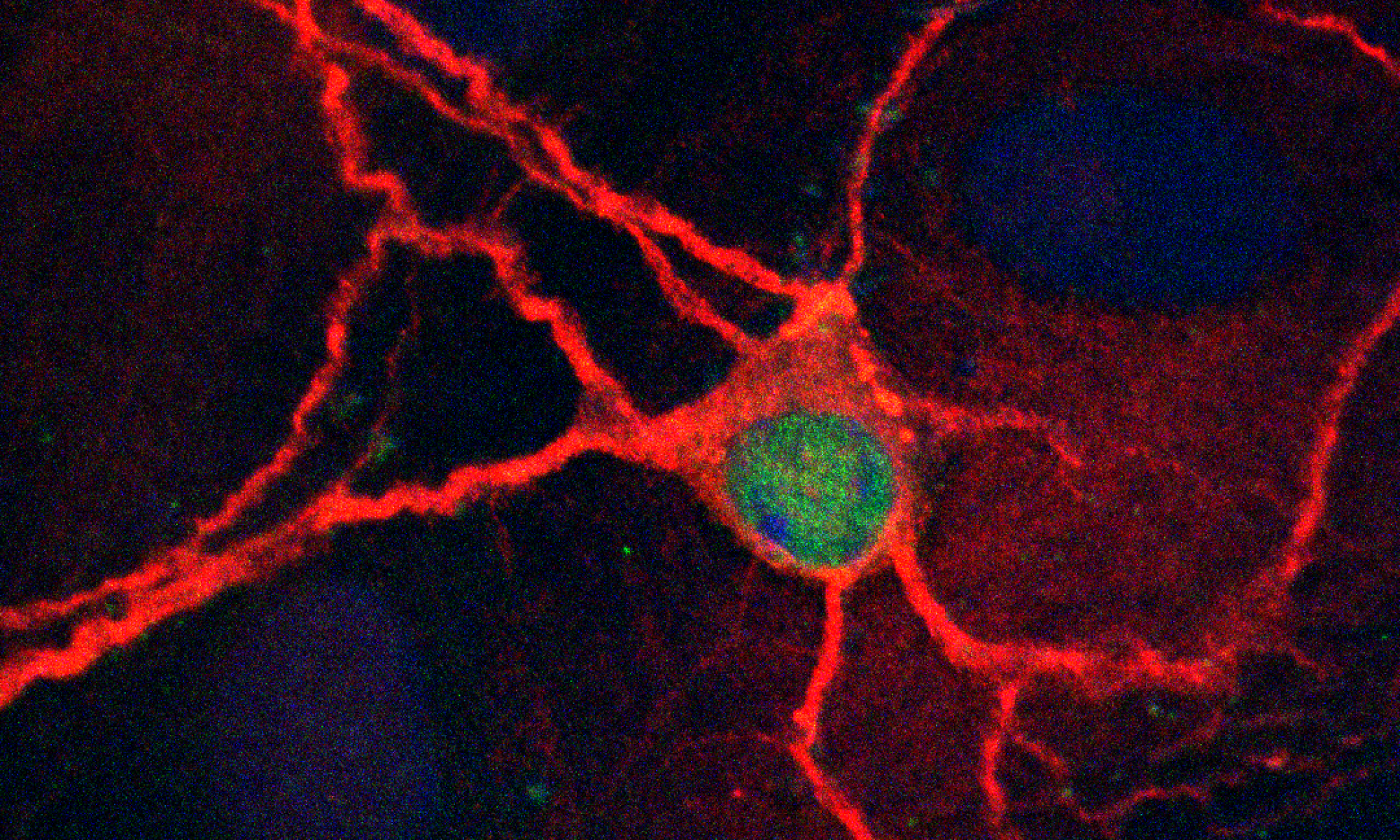
The broad goal of our work is to understand the molecular basis of learning and memory. We began our studies on the protein (FMRP) that is absent in the most common cause of inherited cognitive impairment, fragile X syndrome (FXS). FMRP is an RNA-binding protein that associates with ~4% of brain mRNAs. It has been known for a long time that new protein translation participates in learning and memory. RNA binding proteins like FMRP likely participate in mRNA localization and translation in developing neurons. In addition, microRNAs also participate in these processes and are usually found associated with Argonaute 2 (AGO2). We demonstrated a global role for FMRP in recruiting AGO2 to 3’UTRs. 3’UTRs contain stable guanine-rich structures that form intramolecular G-quadruplexes (GQs). FMRP binds GQs, as does RNA helicase MOV10. We also discovered that FMRP binds MOV10, an AGO2 cofactor. One goal is to understand how FMRP, MOV10, and AGO2 interact with the 3’UTR of co-bound mRNAs to regulate translation to ultimately affect neuronal morphology and function. We are also interested in understanding how MOV10 participates in neuronal development and function.
Exploring the role of MOV10 in brain development
Previously, our lab has demonstrated that MOV10 is highly expressed in the brains of postnatal mice and almost completely absent in the brains of adult mice, suggesting an important developmental role of the protein. Therefore, we created a novel mouse model that has a 90% reduction in MOV10 expression in the excitatory neurons of the brain that we called Mov10 Deletion. We found that the cortical layer II/IV of these mice is thicker than in WT mice, and when we investigated their behavior, we identified that Mov10 Deletion mice have better fear memory. Also, both granule and pyramidal neurons of Mov10 Deletion mice have increased arborization compared to WT. The RNA-seq analysis showed that the absence of MOV10 led to a significant transcriptomic change with a lot of cytoskeletal genes being dysregulated. We then investigated the cytoskeleton dynamics of Mov10 Deletion neurons and found a significantly reduced rate of microtubule nucleation and polymerization. We hypothesized that reduced expression of NuMA1, a protein known to regulate mitotic spindle but also present in dendrites, left unchecked its functional antagonist HAUS protein complex that nucleates new microtubules using existing ones, leading to the observed microtubule phenotype. To test this hypothesis, we overexpressed Numa1 and knocked down Haus6 in Mov10 Deletion neurons and observed a partial rescue effect.
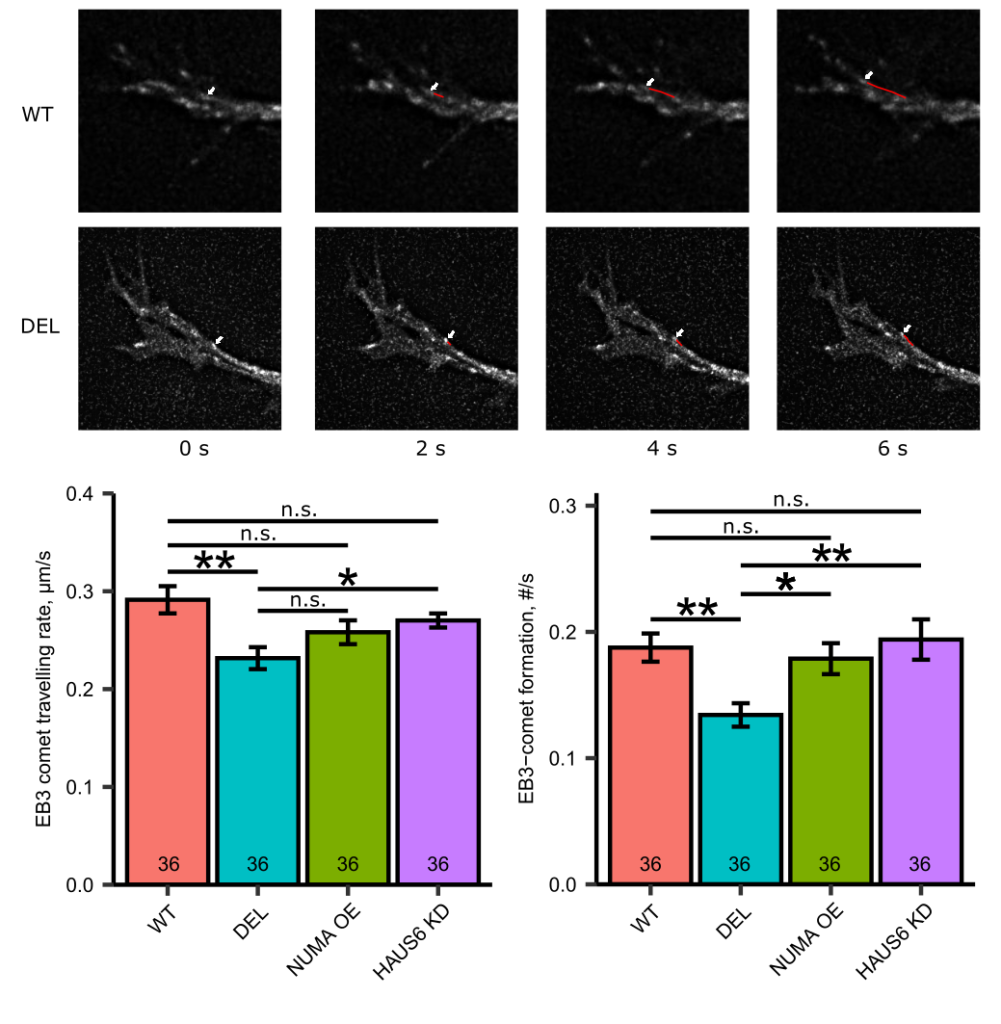
FIGURE: Top panel. Representative images of the EB3-comets in the dendritic cones of DIV2-4 Mov10 Deletion and WT hippocampal neurons over time (seconds). The white arrow shows the path of a single EB3-comet. Bottom opanel. EB3-comet travelling and formation rate in dendritic cones of DIV2 WT and Mov10 Deletion hippocampal neurons transfected with pLenti-CMV-GFP-P2A-Puro (control), pEGFP-P2A-NuMA1, and SMART vector HAUS6 shRNA. Data is shown as mean ± SEM. *p-value <0.05, **p-value < 0.01, n is the number of neurons.
As a continuation of this project, we want to understand if Numa1 mRNA levels are regulated by MOV10 directly or indirectly. We are also interested in understanding the electrophysiology of the Mov10 Deletion neurons since a lot of the differentially expressed genes code for proteins that are important for normal electrophysiological properties of neurons.
Understanding the role of MOV10 in the translational regulation of target RNAs
Bursts of protein translation are required for normal development and neuronal function but it is unknown how this process is regulated. The goal of this research is to explore the key role of RNA unwinding on RNA fate. RNA helicase MOV10 binds in proximity to MREs in 3’UTRs. It unwinds secondary structures in RNA to allow access of the primary effector protein of the miRNA pathway, Argonaute 2, to MREs. By developing an unwinding assay using the iSpinach aptamer, we have demonstrated that MOV10 is able to unwind the most thermodynamically stable RNA secondary structure, the G-Quadruplex (pictured below). Using individual nucleotide Crosslinking Immunoprecipitation (iCLIP), MOV10 was shown to bind mRNAs encoding neuronal projection and cytoskeletal proteins in the developing brain, suggesting a role for MOV10 in regulating translation in dendrites. The hypothesis being tested is that MOV10 binds RNA G-quadruplexes (GQs), which are stable RNA secondary structures, to modulate AGO2 association to MREs in the 3’UTR.
To answer these questions, we will be characterizing a MOV10 conditional knock-out mouse (cKO), identify MOV10 post-translational modifications and how this affects functionality, and characterize the effect MOV10 has on neuronal morphology in situ.
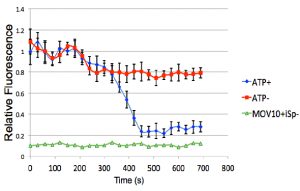
FIGURE: The G-Quadruplex of the iSpinach aptamer is unwound
(measured as a loss in fluorescence) by MOV10 in an
ATP-dependent manner.
(Kenny et al, 2019)