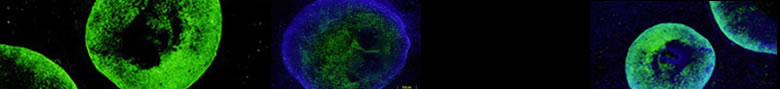
Textbook models of gene activation generally assume a transition from a 30 nm diameter chromatin structure to a beads on a string, 10 nm chromatin fiber during the transition from inactive to active chromatin. We have proceeeded through three generations of experiments on engineered chromosome regions to examine the large-scale chromatin structure of transcriptionally active regions. In all three generations of approaches we observe a consistent decondensation of large-scale chromatin structure accompanying transcriptional activation. However, in contrast to textbook models, we still observe transcription on condensed templates packaged well beyond the 30 nm chromatin fiber. In a 1st generation approach, tethering of transcription activators to a lac operator array in a heterochromatic gene amplified chromosome region was coupled to a dramatic straightening and extension of large-scale chromatin fibers (Figure). A 2nd generation approach instead used cis regulatory elements to activate transcription of a reporter mini-gene contained within plasmid, multi-copy transgene arrays. Large-scale decondensation was less than that observed with the first generation system, either because of use of normal regulatory sequences or due to transgene silencing seen for plasmid arrays which are invariably heterochromatic. In a 3rd generation approach, we observed DHFR, MT, and Hsp70 BAC transgene expression levels within several fold per copy of endogenous genes while packaged in large-scale chromatin fibers condensed well above the 30 nm chromatin fiber (see Commentary). Large-scale chromatin fibers had a linear compaction ratio of ~1000:1 (~25 fold the 40:1 compaction estimated for 30 nm chromatin fibers) decondensing ~1.5-3 fold with transcriptional induction. No evidence for looping of genes outside of these fibers was observed.
Current research is aimed at identifying trans factors mediating large-scale chromatin decondensation associated with transcriptional activation and understanding what the functional implications of large-scale chromatin structure are with regard to transcriptional regulation.
Selected References:
3rd generation approach:
Hu Y, Kireev I, Plutz M, Ashourian N, Belmont AS. Large-scale chromatin structure of inducible genes: transcription on a condensed, linear template. J Cell Biol. 2009 Apr 6; 185 (1) :87-100. PubMed PMID:19349581; PubMed Central PMCID: PMC2700507.
2nd generation approach:
Dietzel S, Zolghadr K, Hepperger C, Belmont AS. Differential large-scale chromatin compaction and intranuclear positioning of transcribed versus non-transcribed transgene arrays containing beta-globin regulatory sequences. J Cell Sci. 2004 Sep 1; 117 (Pt 19) :4603-14. PubMed PMID:15331668.
1st generation approach:
Carpenter AE, Memedula S, Plutz MJ, Belmont AS. Common effects of acidic activators on large-scale chromatin structure and transcription. Mol Cell Biol. 2005 Feb; 25 (3) :958-68. PubMed PMID:15657424; PubMed Central PMCID: PMC544008.
Chen D, Belmont AS, Huang S. Upstream binding factor association induces large-scale chromatin decondensation. Proc Natl Acad Sci U S A. 2004 Oct 19; 101 (42) :15106-11. PubMed PMID:15477594; PubMed Central PMCID: PMC524054.
Carpenter AE, Ashouri A, Belmont AS. Automated microscopy identifies estrogen receptor subdomains with large-scale chromatin structure unfolding activity. Cytometry A. 2004 Apr; 58 (2) :157-66. PubMed PMID:15057969.
Carpenter AE, Belmont AS. Direct visualization of transcription factor-induced chromatin remodeling and cofactor recruitment in vivo. Methods Enzymol. 2004; 375:366-81. PubMed PMID:14870678.
Memedula S, Belmont AS. Sequential recruitment of HAT and SWI/SNF components to condensed chromatin by VP16. Curr Biol. 2003 Feb 4; 13 (3) :241-6. PubMed PMID:12573221.
Nye AC, Rajendran RR, Stenoien DL, Mancini MA, Katzenellenbogen BS, Belmont AS. Alteration of large-scale chromatin structure by estrogen receptor. Mol Cell Biol. 2002 May; 22 (10) :3437-49. PubMed PMID:11971975; PubMed Central PMCID: PMC133805.
Ye Q, Hu YF, Zhong H, Nye AC, Belmont AS, Li R. BRCA1-induced large-scale chromatin unfolding and allele-specific effects of cancer-predisposing mutations. J Cell Biol. 2001 Dec 10; 155 (6) :911-21. PubMed PMID:11739404; PubMed Central PMCID: PMC2150890.
Tumbar T, Sudlow G, Belmont AS. Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J Cell Biol. 1999 Jun 28; 145 (7) :1341-54. PubMed PMID:10385516; PubMed Central PMCID: PMC2133171.