ACES researchers receive University of Illinois racial and social injustice grants


Helping people achieve healthy weight for life
 EMPOWER is an online weight loss program based on previous in-person trials, Individualized Diet Improvement Program (iDip). Moving the program to online greatly increases accessibility and affordability of the program. Furthermore, to improve a success rate of participants, the program is re-designed to address non-dietary issues by collaborating with Dr. Janet Liechty, School of Social Work. Although EMPOWER uses an online platform, it is not an automated program. The core feature is regular evaluation of participants’ progress and provision of advice by a team of professionals in nutrition, social work and medicine. We are currently enrolling Carle patients with obesity-related comorbidities for a clinical trial. More details can be found at EMPOWER.
EMPOWER is an online weight loss program based on previous in-person trials, Individualized Diet Improvement Program (iDip). Moving the program to online greatly increases accessibility and affordability of the program. Furthermore, to improve a success rate of participants, the program is re-designed to address non-dietary issues by collaborating with Dr. Janet Liechty, School of Social Work. Although EMPOWER uses an online platform, it is not an automated program. The core feature is regular evaluation of participants’ progress and provision of advice by a team of professionals in nutrition, social work and medicine. We are currently enrolling Carle patients with obesity-related comorbidities for a clinical trial. More details can be found at EMPOWER.
 MealPlot is a unique mobile app to complement the EMPOWER weight loss program. The main objective of developing this app is to provide the EMPOWER participants with Protein-Fiber Plot at their fingertips, which will enable them to build knowledge and skills to create their own weight loss diets. The app development has been done in collaboration with Applied Research Institute (ARI), College of Engineering. The app can be accessed at mealplot.com.
MealPlot is a unique mobile app to complement the EMPOWER weight loss program. The main objective of developing this app is to provide the EMPOWER participants with Protein-Fiber Plot at their fingertips, which will enable them to build knowledge and skills to create their own weight loss diets. The app development has been done in collaboration with Applied Research Institute (ARI), College of Engineering. The app can be accessed at mealplot.com.
 iDip weight loss trials
iDip weight loss trialsIndividualized Diet Improvement Program (iDip) for sustainable weight loss has been the main project of our lab. The underlying principle of this program is self-empowerment of participants by helping them make an informed decision in food choices. To achieve this goal, the program extensively employs quantitative, visual feedback.
The specific objectives of the project are:
 Achieve safe, 1-2 lb/week weight loss by creating a diet of their liking without relying on dieting products or strict recipes to follow
Achieve safe, 1-2 lb/week weight loss by creating a diet of their liking without relying on dieting products or strict recipes to followLee, M.H., et al., A feasibility study to test a novel approach to dietary weight loss with a focus on assisting informed decision making in food selection. PLOS ONE, 2022. 17(5): p. e0267876.
The Protein-Fiber plot (PF Plot) was developed as a tool for visualizing quantitative nutrient values in foods or meals. A two-dimensional display enables customers to easily compare meals to make an informed choice. When a restaurant menu was displayed as the PF Plot format, customers were able to choose healthier items, whereas providing the same information in numerical tables had no effect at all.
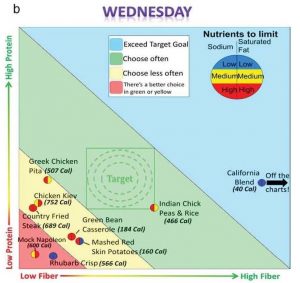 Left: Visualization of nutrient values for easy comparison between foods.
Left: Visualization of nutrient values for easy comparison between foods.
Pratt NS et al. Improvements in recall and food choices using a graphical method to deliver information of select nutrients. Nutr Res 36:44-56, 2016
Our society has made a great progress in reducing diseases caused by nutrient deficiency. However, diseases associated with nutrient excess or imbalances including cardiovascular disease, obesity and diabetes present new challenges to nutrition research. To address these challenges, we need to understand how our body adjusts its metabolism to a wide variety of diets.
A central mechanism for this adaptive response is changing the expression of genes involved in the metabolic process. Our group has been investigating molecular mechanisms by which macronutrients (protein, carbohydrate and fat) and their metabolites regulate gene expression. Advancing our knowledge of the fundamental mechanism underlying metabolic adaptation, or failure of adaptation, will greatly contribute to the development of efficacious strategies for the dietary prevention/treatment of obesity and chronic metabolic diseases.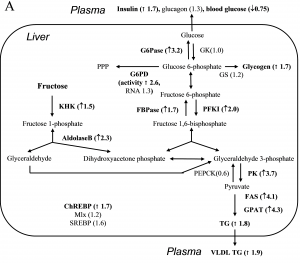
Right: Induction of genes by dietary fructose.
Koo HY et al. Dietary fructose induces a wide range of genes with distinct shift in carbohydrate and lipid metabolism in fed and fasted rat liver. Biochim Biophys Acta 1782:341-348, 2008