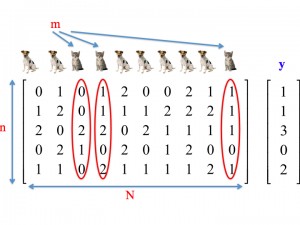
Correlation Clustering
Closely related to rank aggregation and other problems that fall within the umbrella of optimization on graphs. We considered the problem of correlation clustering on graphs with constraints on both the cluster sizes and the positive and negative weights of edges. Our contributions include introducing the problem of correlation clustering with bounded cluster sizes; Second, extending the region of weight values for which the clustering may be performed with constant approximation guarantees in polynomial time and apply the results to the bounded cluster size problem.
In collaboration with J. Ma, we are also working on applying our correlation clustering algorithms to problems in cancer genomics and cancer pathway analysis.
Ordinal data processing
Ordinal data (ranking or preference data) is an emerging representation format in social sciences, recommender systems, rank modulation for flash memories and crowdvoting system.
From the viewpoint of signal processing and information theory, this data structure has been largely overlooked and many problems regarding ordinal data mining, learning, compression, synchronization remain unsolved.
Our work covers four important aspects of ordinal data processing
1) Rank comparison and rank aggregation, with application to search, machine learning and bioinformatics.
2) Learning how to rank and compressing rankings, with applications to data storage, modeling and data mining.
3) Synchronizing ordinal data, for playlist, tasklist and search query reconciliation.
4) Error-correction codes for ordinal data, with special emphasis on the Ulam metric.
Collaborators on these topics include Farzad Farnoud, Hanmao Kiah, Minji Kim, Arya Mazumdar and Fardad Raisali.
Data Deduplication and Synchronization
Due to large data volumes and networked user access, addressing the problems of deduplicating and synchronizing data in distributed storage systems is gaining increasing interest. Our work is focusing on deduplication and synchronization under edits that include deletions and insertions which operate on coded data. Recent results include modifications of MDS, regenerating and locally repairable codes that allow updates in the parity-check values to be performed with one round of communication at low bit rates and using small storage overhead. Our main contributions to this area are novel protocols for synchronizing both frequently updated and semi-static data and protocols for data deduplication applications, based on intermediary permutation, Vandermonde and Cauchy matrix coding.
Group Testing
Group testing is a combinatorial pooling techniques that exploits sparsity of subjects with negative traits to reduce the number of tests needed for identification. It has a long history, dating back to the early 1940’s and pooling soldiers for STDs in WWII.
Nowadays, pooling is frequently used in genomic screening and genotyping application.
Our work is centered around a new testing paradigm that addresses issues that arise in practical biotechnology applications. The underlying paradigm is referred to semiquantitative group testing and it may be viewed as a general concatenation of adder models and quantizers.
This is joint work with Amin Emad.
DNA-Based Storage
DNA based storage has attracted significant attention due to recent
demonstrations of the viability of storing information in macromolecules.
Unlike classical optical and magnetic storage technologies, DNA based
storage does not require electrical supply to maintain data integrity,
and given the trends in cost decreases of DNA synthesis and sequencing,
it is now widely acknowledged that within the next $10-15$ years
DNA storage may represent the predominant archiving technology; currently,
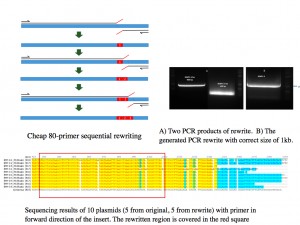
there are no known solutions for rewritable and random access DNA
based storage. Our work is the first step in the direction of designing functional
rewritable DNA-based storage systems based on sophisticated coding approaches and
synthetic biology.
This is joint work with Yong Bo, Jian Ma, Hussein Tabatabataie and Huimin Zhao.
Genomic Data Compression
With the rapid development of modern sequencing technologies, the volumes of genomic data have experienced tremendous growths, leading to the need for developing specialized compression techniques suitable for DNA sequence reads. Our work focuses both on de novo and reference based compression methods for metagenomic and RNA-Seq data.
This is joint work with Minji Kim, Zhying Wang and Tsachy Weissmann.
In addition, our recent work includes developing new reverse-engineering methods for gene regulatory networks based on compressive sensing techniques coupled with causal inference; synchronization for distributed storage codes; string reconstruction from traces and others.
PRIOR WORK
For our work in constrained coding, combinatorial urn model analysis, LDPC and algebraic codes, compressive sensing and low-rank matrix completion, please check the full list of publications.