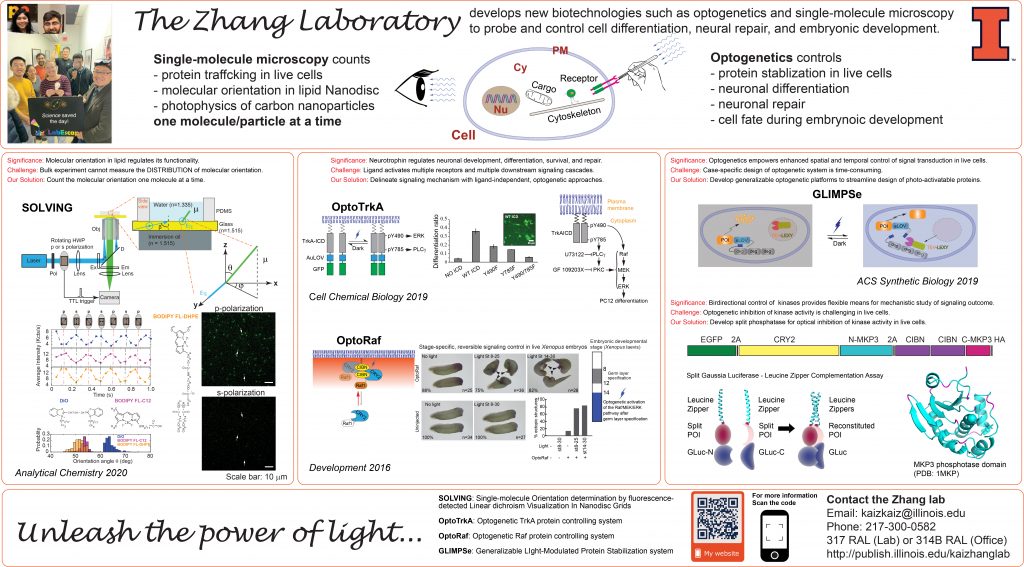
Here is a summary of our research that I have presented at the recent 2021 PacificChem conference. Hope you enjoy it.
Synthetic Biology – Control Molecular Activity and Steering Cell Fate with Optogenetics
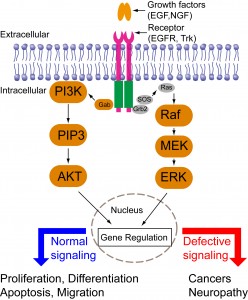
Cells are constantly making decisions in response to their environments. Intracellular signal transduction transmits external stimuli into the cell interior and regulates transcription and translation. Growth factor-mediated signal transduction regulates a wide spectrum of cellular functions such as cell proliferation, differentiation, migration, and apoptosis (Fig. 1). Dysregulated growth factor signaling has been observed in various diseases including cancers or neurological disorders. Intriguingly, different growth factors trigger distinct cellular functions via activation of similar downstream signaling cascades such as the mitogen-activated protein kinase (MAPK), phosphoinositide 3-kinase (PI3K)-AKT, and phospholipase C (PLC). Consequently, a central question in growth-factor mediated signal transduction is how the same set of signaling cascades elicits such diverse yet specific cellular outcomes. Previous research has suggested that cells employ spatiotemporal regulation of their signaling cascades to convey specificity of cell fate determination. For instance, epidermal growth factor (EGF) triggers a transient MAPK activation and induces cell proliferation; nerve growth factor (NGF) triggers a sustained MAPK activation and induces cell differentiation. Directed cell migration is driven by asymmetric subcellular activation of actin dynamics. Conventional pharmacological and genetic approaches have continuously expanded our knowledge base of signaling components involved in signaling networks. These approaches, however, lack the resolution of spatial and temporal control. A better understanding of signaling mechanisms, therefore, calls for new tools that can precisely control intracellular signaling in both space and time.
Optogenetics combines the power of light and genetics and enables precise spatial and temporal control of individual signaling cascades. Our previous work used light to control the MAPK signaling pathway and quantitatively revealed the kinetic effect of the MAPK signaling on cell differentiation (Mov. 1 and 2). In the long term, we aim to 1) extend optogenetic modules for activating other growth factor signaling pathways, 2) dissect how spatiotemporal regulation of growth-factor signal pathways determine cell fate, and 3) develop new optogenetic tools using protein engineering, computation, and genetic screening.
Recent publications:
52. Qin Wang, Huaxun Fan, Feng Li, Savanna R. Sharum, Vishnu Krishnamurthy, Yuanquan Song*, Kai Zhang* “Spatiotemporal optogenetic control of neurotrophin signaling accelerates axon regeneration and functional recovery in the peripheral and central nervous system”, eLife, 2020, 9: e57395 [Link][PDF] [bioRxiv].
47. Vishnu V. Krishnamurthy, Jia Fu, Teak-Jung Oh, John Khamo, Jing Yang*, Kai Zhang* “A Generalizable Optogenetic Strategy to Regulate Receptor Tyrosine Kinases during Vertebrate Embryonic Development”, Journal of Molecular Biology, 2020, 432, 10, 3149-3158. https://doi.org/10.1016/j.jmb.2020.03.032 [Link][PDF].
37. John S. Khamo, Vishnu V. Krishnamurthy, Qixin Chen, Jiajie Diao*, and Kai Zhang*, “, Cell Chemical Biology, 2018 DOI: https://doi.org/10.1016/j.chembiol.2018.11.004 [Link][PDF].
32. John S. Khamo, Vishnu V. Krishnamurthy, Payel Mondal, Savanna R. Sharum, and Kai Zhang* “Applications of optobiology in intact cells and multi-cellular organisms”, Journal of Molecular Biology, 2017, 429, 2999-3017. (DOI: 10.1016/j.jmb.2017.08.015)[Link][PDF]
27. V. V. Krishnamurthy, J.S. Khamo, W. Mei, A. J. Turgeon, H. M. Ashraf, P. Mondal, D. B. Patel, N. Risner, E. E. Cho, J. Yang*, and K. Zhang* “Reversible optogenetic control of kinase activity during differentiation and embryonic development” Development, 2016, 143, 4085-4094. (DOI: 10.1242/dev.140889) [Link][PDF]
Figure 1. Signaling pathways downstream of growth factors binding to membrane receptors.
Movie 1. Light-induced ERK nuclear translocation.
Movie 2. Reversible ERK activation in live cells.
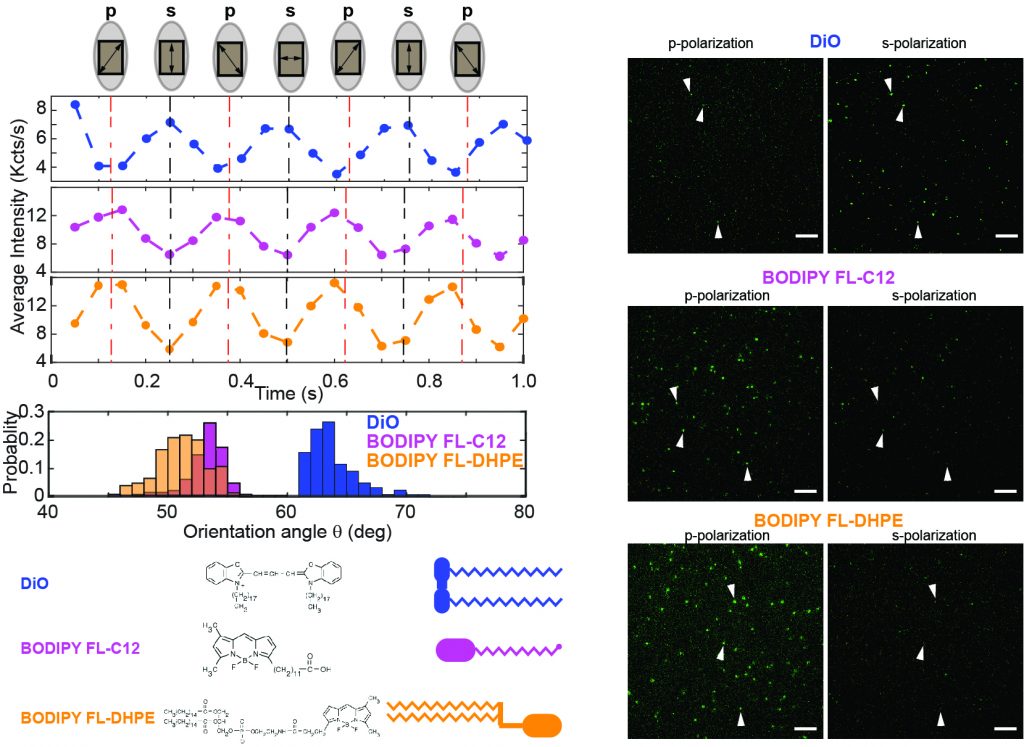
Resolving the molecular orientation inside single Nanodiscs
The function of membrane-bound proteins often depends on their interactions with the lipid bilayer. Bulk absorption-based linear dichroism has been historically used to investigate molecular orientations in the phospholipid bilayer but cannot resolve the actual distribution of molecules embedded in the membrane and is often limited by a poor signal-to-noise ratio. Here, we present single-molecule orientation determination by fluorescence-detected linear dichroism visualization in Nanodisc grids or SOLVING, to determine the molecular orientation of molecules assembled into nanoscale lipid bilayers. SOLVING provides the actual distribution of orientations and promises to provide key molecular insights into the topology and interactions of multiprotein complexes, such as those observed in intracellular signal transduction.
Recent publications:
45. Tyler Camp, Kritika Mehta, Stephen Sligar*, Kai Zhang*, “Molecular Orientation Determination in Nanodiscs at the Single-Molecule Level”, Analytical Chemistry, 2019, 92, 2, 2229-2236 [Link][PDF].
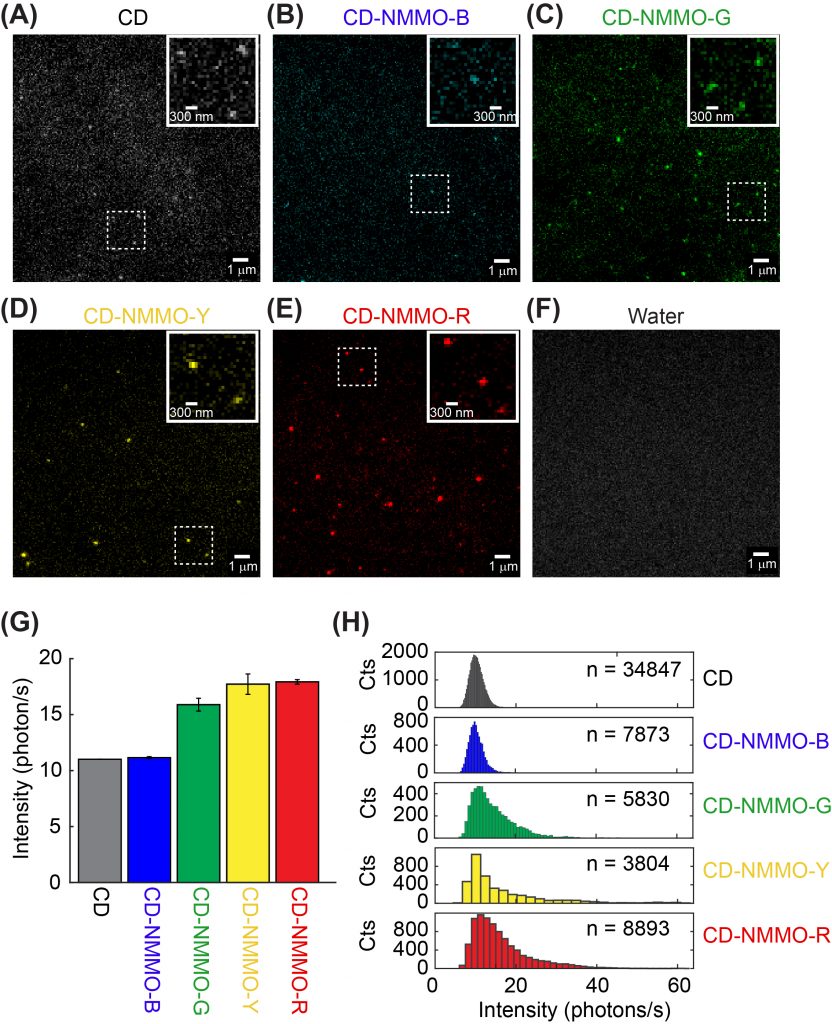
Photophysics of fluorescent carbon nanodots revealed at the single-particle level
Fluorescent carbon dots are emerging nanometer-sized materials that are blessed with biocompatibility, versatile emission wavelength tuning, photostability, and economical synthesis. Recently, fluorescent carbon dots have been increasingly used in bioimaging and biosensing, although their photophysics has not been well understood. Unlike our previously investigated nanoparticles such as quantum dots, photophysics of carbon dots depends on their surface states instead of particle size. In a collaboration with Dr. Dipanjan Pan, we set out to investigate the photophysics of carbon dots at the single-particle level. We are particularly interested in determining how synthetic conditions such as temperature and precursor composition affects the fluorescent emission properties of carbon dots.
Recent publications:
43. Indrajit Srivastava, John S. Khamo, Subhendu Pandit, Parinaz Fathi, Xuedong Huang, Anleen Cao, Richard T. Haasch, Shuming Nie, Kai Zhang*, Dipanjan Pan* “Influence of Electron Acceptor and Electron Donor on the Photophysical Properties of Carbon Dots: A Comparative Investigation at the Bulk‐State and Single‐Particle Level. Advanced Functional Materials, 2019, 1902466 [Link][PDF].
38. Parinaz Fathi, John S. Khamo, Xuedong Huang, Indrajit Srivastava, Mandy B. Esch, Kai Zhang*, and Dipanjan Pan* “Bulk-state and single-particle imaging are central to understanding carbon dot photo-physics and elucidating the effects of precursor composition and reaction temperature” Carbon, 2019, 145, 572-585. [Link] [PDF].
35. Santosh K. Misra, Indrajit Srivastava, John S. Khamo, Vishnu V. Krishnamurthy, Dinabandhu Sar, Aaron S. Schwartz-Duval, Julio A. N. T. Soares, Kai Zhang*, and Dipanjan Pan* “Carbon Dots with Induced Surface Oxidation Permits Imaging at Single-Particle Level for Intracellular Studies”, Nanoscale, 2018, 10(39) 18510-18519. (DOI: 10.1039/c8nr04065f). [Link][PDF]
Axonal transport in neurological disorders
Neurons are the most polarized cell types, extending processes 10,000 times the size of their cell bodies. In such a polarized cell, Brownian diffusion is not sufficient to drive efficient communications across the whole neuron cell. For a small molecule to diffuse from the hand to the brain, it will take about 30 years! Instead, communication between different parts of neuronal cells requires active transport. Cargoes containing signaling molecules or newly synthesized proteins are transported along the cytoskeletal tracks of axons, very similar to the vehicle transport on highways. Defective axonal transport has been observed in neurological disorders such as Alzheimer’s disease, Parkinson’s disease, Amyotrophic Lateral Sclerosis (ALS), and Charcot-Marie-Tooth neuropathy, to name a few.
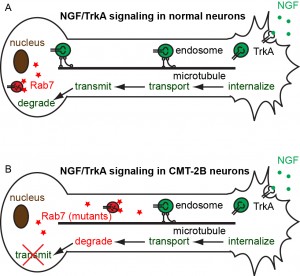
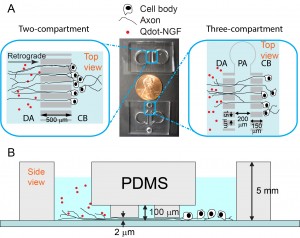
The Zhang group studies the neurotrophic signaling pathway that primarily regulates neuronal differentiation and survival. Our previous work has shown that axonal transport can be either slowed down or accelerated in neurological disorders (Fig. 3). We use compartmentalized microfluidic devices to spatially segregate and control the chemical environment of axons and cell bodies (Fig. 4). We use live cell imaging to track the axonal transport of fluorescently labeled cargos. Our long term goal is to determine how axonal transport is affected by specific neuronal phenotypes and how manipulation of axonal transport may rescue these phenotypes.
Figure 3. Model of defective axonal transport of Rab7 in Charcot-Marie-Tooth 2B neuropathy.
Figure 4. Microfluidic devices for live cell tracking of axonal transport.
Movie 3. Axonal transport of TrkA-mCherry in dorsal root ganglion neurons cultured in microfluidic channels.
Recent publications:
33. , , , , , , , , , , , , and
30. Y. Osakada, K. Zhang “Single-particle tracking reveals a dynamic role of actin filaments in assisting long-range axonal transport in neurons” Bulletin of the Chemical Society of Japan (BCSJ), 2017, 90, 714-719. (DOI: 10.1246/bcsj.20170090) [Link][PDF]
Super-resolution imaging of in live cells
Recent publications:
40. Qixin Chen, Xintian Shao, Zhiqi Tian, Yang Chen, Payel Mondal, Fei Liu, Fengshan Wang, Peixue Ling*, Weijiang He*, Kai Zhang*, Zijian Guo, and Jiajie Diao*, “Nanoscale Monitoring of Mitochondria and Lysosome Interactions for Drug Screening and Discovery”, Nano Research, 2019, 12, 5, 1009–1015. [Link][PDF].
39. Bin Cai, Luning Yu, Savanna R. Sharum, Kai Zhang*, and Jiajie Diao*, “Single-vesicle measurement of protein-induced membrane tethering”, Colloids and Surfaces B: Biointerfaces, 2019, 177, 267-273. [Link][PDF].
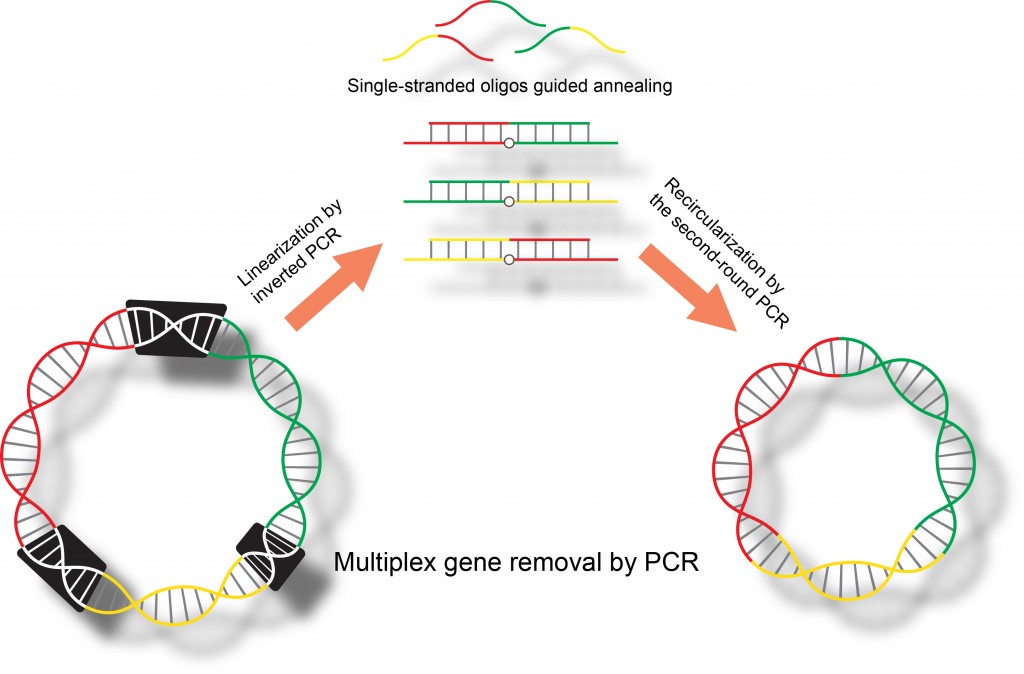
Genetic engineering for synthetic biology
Recent publications:
28. Vishnu V. Krishnamurthy, and Kai Zhang* ” Simultaneous removal of multiple DNA segments by polymerase chain reactions” Methods in Molecular Biology, Synthetic DNA, Ed R. Hughes. (Springer New York) 2017, 1472, 193-203. (DOI: 10.1007/978-1-4939-6343-0_15) [Link][PDF]
25. Vishnu V. Krishnamurthy, John S. Khamo, Ellen Cho, Cara Schornak, and Kai Zhang* “Multiplex gene removal by two-step polymerase chain reactions”, Analytical Biochemistry, 2015, 481, 7-9. (DOI: 10.1016/j.ab.2015.03.033). [Link][PDF]